6V6P
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
4WRT
 
 | | Crystal structure of Influenza B polymerase with bound vRNA promoter (form FluB2) | | Descriptor: | Influenza virus polymerase vRNA promoter 3' end, Influenza virus polymerase vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-25 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
6LQ0
 
 | |
6EHB
 
 | |
6LKM
 
 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 5-chloro-N-((1S,2R)-2-(6-fluoro-2,3-dimethylphenyl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)-4-methyl-3,4-dihydro-2H-benzo[b][1,4]oxazine-8-sulfonamide | | Descriptor: | 5-chloro-N-((1S,2R)-2-(6-fluoro-2,3-dimethylphenyl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)-4-methyl-3,4-dihydro-2H-benzo[b][1,4]oxazine-8-sulfonamide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
8SDI
 
 | |
8SDE
 
 | |
8FHU
 
 | |
8FFY
 
 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(UGA-TL) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Serine--tRNA ligase, mitochondrial, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-12-11 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for a degenerate tRNA identity code and the evolution of bimodal specificity in human mitochondrial tRNA recognition.
Nat Commun, 14, 2023
|
|
5L7X
 
 | | Afamin antibody fragment, N14 Fab, L1- glycosylated, crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Mouse Antibody Fab Fragment, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8RXR
 
 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | Descriptor: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 18, 2024
|
|
7L6M
 
 | |
7KS9
 
 | |
8HXE
 
 | | Crystal structure of B3 L1 MBL in complex with 2-amino-5-(4-propylbenzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-propylphenyl)methyl]-1,3-thiazole-4-carboxylic acid, Metallo-beta-lactamase L1 type 3, THIOCYANATE ION, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-04 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
7L3N
 
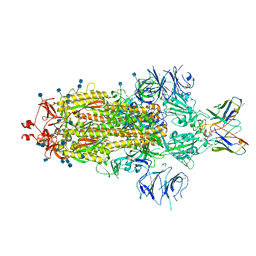 | | SARS-CoV 2 Spike Protein bound to LY-CoV555 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV555 Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection.
Biorxiv, 2020
|
|
5KKE
 
 | |
2RTD
 
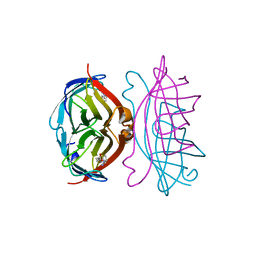 | | STREPTAVIDIN-BIOTIN COMPLEX, PH 1.39, SPACE GROUP I222 | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
8FF7
 
 | |
8HXI
 
 | | Crystal structure of B3 L1 MBL in complex with 2-amino-5-(4-isopropylbenzyl)thiazole-4-carboxylic acid | | Descriptor: | 2-azanyl-5-[(4-propan-2-ylphenyl)methyl]-1,3-thiazole-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase L1 type 3, ... | | Authors: | Yan, Y.-H, Zhu, K.-R, Li, G.-B. | | Deposit date: | 2023-01-04 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of 2-Aminothiazole-4-carboxylic Acids as Broad-Spectrum Metallo-beta-lactamase Inhibitors by Mimicking Carbapenem Hydrolysate Binding.
J.Med.Chem., 66, 2023
|
|
8FF6
 
 | | Cytosolic ascorbate peroxidase mutant from Panicum virgatum | | Descriptor: | Cytosolic ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Activity of Cytosolic Ascorbate Peroxidase (APX) from Panicum virgatum against Ascorbate and Phenylpropanoids.
Int J Mol Sci, 24, 2023
|
|
8FNW
 
 | | Structure of RdrA-RdrB complex from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, Archaeal ATPase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNT
 
 | | Structure of RdrA from Escherichia coli RADAR defense system | | Descriptor: | Archaeal ATPase | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
1BGO
 
 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PEPTIDOMIMETIC INHIBITOR | | Descriptor: | 1-[2-(3-BIPHENYL)-4-METHYLVALERYL)]AMINO-3-(2-PYRIDYLSULFONYL)AMINO-2-PROPANONE, CATHEPSIN K | | Authors: | Desjarlais, R.L, Yamashita, D.S, Oh, H.-J, Bondinell, W.E, Uzinskas, I.N, Erhard, K.F, Allen, A.C, Haltiwanger, R.C, Zhao, B, Smith, W.W, Abdel-Meguid, S.S, D'Alessio, K, Janson, C.A, Mcqueney, M.S, Tomaszek, T.A, Levy, M.A, Veber, D.F. | | Deposit date: | 1998-05-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of X-Ray Co-Crystal Structures and Molecular Modeling to Design Potent and Selective Non-Peptide Inhibitors of Cathepsin K
J.Am.Chem.Soc., 120, 1998
|
|
7ZNJ
 
 | | Structure of an ALYREF-exon junction complex hexamer | | Descriptor: | Eukaryotic initiation factor 4A-III, N-terminally processed, MAGNESIUM ION, ... | | Authors: | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | Deposit date: | 2022-04-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
