3PD6
 
 | | Crystal structure of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV.
Biosci.Rep., 31, 2011
|
|
3PDB
 
 | | Crystal structure of mouse mitochondrial aspartate aminotransferase in complex with oxaloacetic acid | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, mitochondrial, ... | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV.
Biosci.Rep., 31, 2011
|
|
8SKR
 
 | | human liver mitochondrial Aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhang, Z, Tringides, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
9AAT
 
 | |
6AY8
 
 | |
6AY7
 
 | | Cartilage homing cysteine-dense-peptides | | Descriptor: | CHLORIDE ION, Potassium channel toxin alpha-KTx 3.5, SULFATE ION, ... | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2017-09-07 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cartilage homing cysteine-dense-peptides
To Be Published
|
|
1F1G
 
 | | Crystal structure of yeast cuznsod exposed to nitric oxide | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, PHOSPHATE ION, ... | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-12 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F18
 
 | | Crystal structure of yeast copper-zinc superoxide dismutase mutant GLY85ARG | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F1A
 
 | | Crystal structure of yeast H48Q cuznsod fals mutant analog | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F1D
 
 | | Crystal structure of yeast H46C cuznsod mutant | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
8BAH
 
 | | Human Mre11-Nbs1 complex | | Descriptor: | Double-strand break repair protein MRE11, MANGANESE (II) ION, Nibrin | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-10-11 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
6SQP
 
 | | Crystal structure of Cat MDM2-S429E RING domain homodimer | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, NITRATE ION, ... | | Authors: | Magnussen, H.M, Ahmed, S.F, Huang, D.T. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain.
Nat Commun, 11, 2020
|
|
1N5Y
 
 | | HIV-1 Reverse Transcriptase Crosslinked to Post-Translocation AZTMP-Terminated DNA (Complex P) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3', 5'-D(*AP*TP*GP*C*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, P, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 Reverse Transcriptase with Pre-Translocation and Post-Translocation AZTMP-Terminated DNA
Embo J., 21, 2002
|
|
1N6Q
 
 | | HIV-1 Reverse Transcriptase Crosslinked to pre-translocation AZTMP-terminated DNA (complex N) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3', 5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, I, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-11 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 Reverse Transcriptase with Pre- and Post-translocation AZTMP-terminated DNA
Embo J., 21, 2002
|
|
3V81
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and the nonnucleoside inhibitor nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8503 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V6D
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) cross-linked with AZT-terminated DNA | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7048 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V4I
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and AZTTP | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7983 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2DKI
 
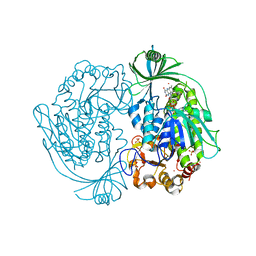 | | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni, under pressure of xenon gas (12 atm) | | Descriptor: | 3-HYDROXYBENZOATE HYDROXYLASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hiromoto, T, Fujiwara, S, Hosokawa, K, Yamaguchi, H. | | Deposit date: | 2006-04-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site
J.Mol.Biol., 364, 2006
|
|
1GXC
 
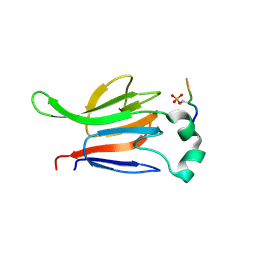 | | FHA domain from human Chk2 kinase in complex with a synthetic phosphopeptide | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE CHK2, SYNTHETIC PHOSPHOPEPTIDE | | Authors: | Li, J, Williams, B.L, Haire, L.F, Goldberg, M, Wilker, E, Durocher, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-13 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Versatility of the Fha Domain in DNA-Damage Signaling by the Tumor Suppressor Kinase Chk2
Mol.Cell, 9, 2002
|
|
5MNJ
 
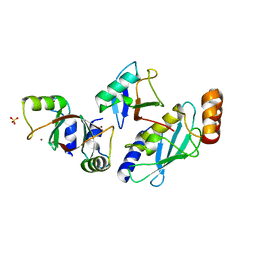 | | Structure of MDM2-MDMX-UbcH5B-ubiquitin complex | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Polyubiquitin-B, Protein Mdm4, ... | | Authors: | Klejnot, M, Huang, D.T. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural analysis of MDM2 RING separates degradation from regulation of p53 transcription activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3TF1
 
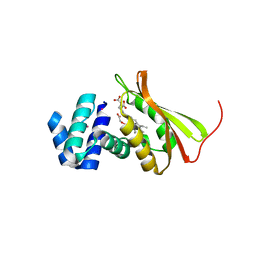 | | Crystal structure of an H-NOX protein from T. tengcongensis under 6 atm of xenon | | Descriptor: | ACETATE ION, Methyl-accepting chemotaxis protein, OXYGEN MOLECULE, ... | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0369 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TFE
 
 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120, L66W mutant under 6 atm of xenon | | Descriptor: | Alr2278 protein, MALONIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SZM
 
 | |
3TFA
 
 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120 under 6 atm of xenon | | Descriptor: | Alr2278 protein, PROTOPORPHYRIN IX CONTAINING FE, XENON | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2711 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TF9
 
 | | Crystal structure of an H-NOX protein from Nostoc sp. PCC 7120 under 1 atm of xenon | | Descriptor: | Alr2278 protein, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Winter, M.B, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5901 Å) | | Cite: | Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
