8FID
 
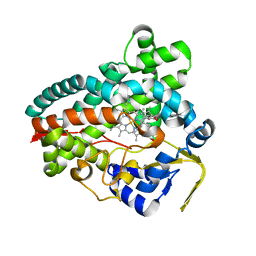 | | Crystal Structure of Erwinia tracheiphila CYP114 in complex with ent-kaurenoic acid (Crystal Form 2) | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-kaur-16-en-18-oic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Stewart Jr, C.E, Nagel, R. | | Deposit date: | 2022-12-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Dual factors required for cytochrome-P450-mediated hydrocarbon ring contraction in bacterial gibberellin phytohormone biosynthesis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FY7
 
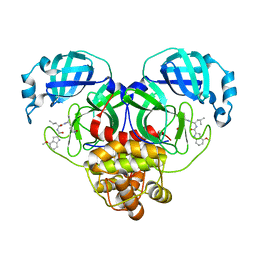 | | SARS-CoV-2 main protease in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase nsp5, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-1-[(3S)-2-oxopyrrolidin-3-yl]but-3-en-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Fried, W, Chen, X.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
8FAI
 
 | | Cryo-EM structure of the Agrobacterium T-pilus | | Descriptor: | (7Z,19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacos-7-en-19-yl (9Z)-octadec-9-enoate, Protein virB2 | | Authors: | Kreida, S, Narita, A, Johnson, M.D, Tocheva, E.I, Das, A, Jensen, G.J, Ghosal, D. | | Deposit date: | 2022-11-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the Agrobacterium tumefaciens T4SS-associated T-pilus reveals stoichiometric protein-phospholipid assembly.
Structure, 31, 2023
|
|
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8FIC
 
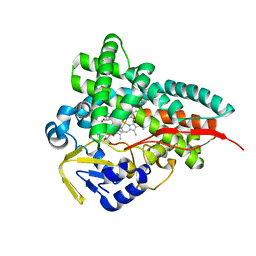 | | Crystal Structure of Erwinia tracheiphila CYP114 in complex with ent-kaurenoic acid (Crystal Form 1) | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-kaur-16-en-18-oic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Stewart Jr, C.E, Nagel, R. | | Deposit date: | 2022-12-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dual factors required for cytochrome-P450-mediated hydrocarbon ring contraction in bacterial gibberellin phytohormone biosynthesis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GK3
 
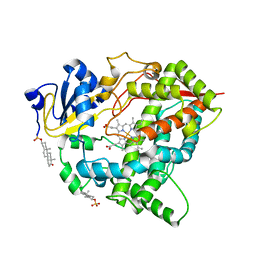 | | Cytochrome P450 3A7 in complex with Dehydroepiandrosterone sulfate | | Descriptor: | 17-oxoandrost-5-en-3beta-yl hydrogen sulfate, Cytochrome P450 3A7, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, J, Scott, E.E. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human cytochrome P450 3A7 binding four copies of its native substrate dehydroepiandrosterone 3-sulfate.
J.Biol.Chem., 299, 2023
|
|
8GLE
 
 | | Crystal Structure of Human CD1b in Complex with Lysosulfatide | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl 3-O-sulfo-beta-D-galactopyranoside, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shahine, A. | | Deposit date: | 2023-03-22 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms.
Cell, 186, 2023
|
|
8G6F
 
 | | Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, Proteasome endopeptidase complex, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
8SMR
 
 | | cytochrome bc1-cbb3 supercomplex from Pseudomonas aeruginosa | | Descriptor: | (2R)-2-(methoxymethyl)-4-{[(25R)-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Di Trani, J.M, Rubinstein, J.L. | | Deposit date: | 2023-04-26 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the bc 1 - cbb 3 respiratory supercomplex from Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SNH
 
 | | cytochrome bc1-cbb3 supercomplex from Pseudomonas aeruginosa | | Descriptor: | (2R)-2-(methoxymethyl)-4-{[(25R)-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Di Trani, J.M, Rubinstein, J.L. | | Deposit date: | 2023-04-27 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the bc 1 - cbb 3 respiratory supercomplex from Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T6L
 
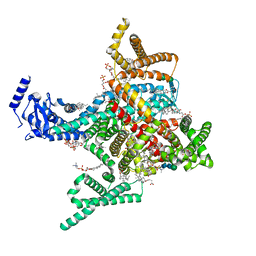 | | Cryo-EM structure of rat cardiac sodium channel NaV1.5 with batrachotoxin analog BTX-B | | Descriptor: | (1R)-1-[(5aR,7aR,9R,11aS,11bS,12R,13aR)-9,12-dihydroxy-2,11a-dimethyl-1,2,3,4,7a,8,9,10,11,11a,12,13-dodecahydro-7H-9,11b-epoxy-13a,5a-prop[1]enophenanthro[2,1-f][1,4]oxazepin-14-yl]ethyl benzoate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Tonggu, L, Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dual receptor-sites reveal the structural basis for hyperactivation of sodium channels by poison-dart toxin batrachotoxin.
Nat Commun, 15, 2024
|
|
8SSI
 
 | |
8T4Z
 
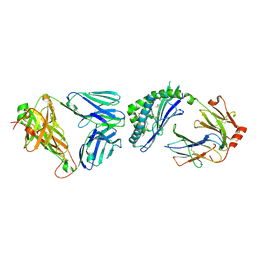 | | T-cell receptor and lipid complex structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, Beta-2-microglobulin, ... | | Authors: | Praveena, T, Rossjohn, J. | | Deposit date: | 2023-06-12 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Lipidomic scanning of self-lipids identifies headless antigens for natural killer T cells.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ST7
 
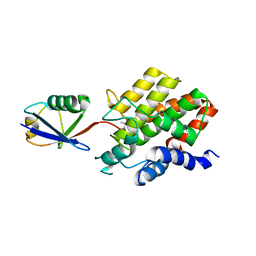 | | Structure of E3 ligase VsHECT bound to ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase SopA-like catalytic domain-containing protein, Ubiquitin, prop-2-en-1-amine | | Authors: | Franklin, T.G, Pruneda, J.N. | | Deposit date: | 2023-05-09 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Bacterial ligases reveal fundamental principles of polyubiquitin specificity.
Mol.Cell, 83, 2023
|
|
8ST9
 
 | |
8ST8
 
 | |
8STH
 
 | | human STING with diABZI agonist 15 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-(3-hydroxypropoxy)-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Duvall, J.R, Bukhalid, R.A. | | Deposit date: | 2023-05-10 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Optimization of a STING Agonist Platform for Application in Antibody Drug Conjugates.
J.Med.Chem., 66, 2023
|
|
8STI
 
 | | human STING with agonist XMT-1616 | | Descriptor: | 3-[(2E)-4-{5-carbamoyl-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-(3-hydroxypropoxy)-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-3H-imidazo[4,5-b]pyridine-6-carboxamide, Stimulator of interferon genes protein | | Authors: | Duvall, J.R, Bukhalid, R.A. | | Deposit date: | 2023-05-10 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and Optimization of a STING Agonist Platform for Application in Antibody Drug Conjugates.
J.Med.Chem., 66, 2023
|
|
8R67
 
 | | tubulin-cryptophycin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(3~{S},10~{R},13~{E},16~{S})-10-[(3-chloranyl-4-methoxy-phenyl)methyl]-6,6-dimethyl-2,5,9,12-tetrakis(oxidanylidene)-16-[(1~{S})-1-[(2~{R},3~{R})-3-phenyloxiran-2-yl]ethyl]-1,4-dioxa-8,11-diazacyclohexadec-13-en-3-yl]ethanoic acid, CALCIUM ION, ... | | Authors: | Abel, A.C, Muehlethaler, T, Dessin, C, Steinmetz, M.O, Sewald, N, Prota, A.E. | | Deposit date: | 2023-11-21 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bridging the maytansine and vinca sites: Cryptophycins target beta-tubulin's T5-loop.
J.Biol.Chem., 300, 2024
|
|
8TWY
 
 | | Structure of PIK3CA with covalent inhibitor ALO26 | | Descriptor: | 1-(4-{[2-(4-{(4P)-4-[2-amino-4-(difluoromethyl)pyrimidin-5-yl]-6-[(3S)-3-methylmorpholin-4-yl]-1,3,5-triazin-2-yl}piperazin-1-yl)-2-oxoethoxy]methyl}piperidin-1-yl)prop-2-en-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, Barlow-Busch, I. | | Deposit date: | 2023-08-21 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure of PIK3CA with covalent inhibitor ALO26
To Be Published
|
|
8V4P
 
 | |
8V15
 
 | | Human SIRT3 bound to p53-AMC peptide, Carba-NAD, and Honokiol | | Descriptor: | (1P)-3',5-di(prop-2-en-1-yl)[1,1'-biphenyl]-2,4'-diol, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, ... | | Authors: | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | Deposit date: | 2023-11-19 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8VC1
 
 | | CryoEM structure of insect gustatory receptor BmGr9 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, ... | | Authors: | Frank, H.M, Walsh Jr, R.M, Garrity, P.A, Gaudet, R. | | Deposit date: | 2023-12-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure of an insect gustatory receptor.
Biorxiv, 2023
|
|
8SX8
 
 | |
8SX7
 
 | |
