6RZG
 
 | | Galectin-3C in complex with meta-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.015 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
3L7M
 
 | |
6RQB
 
 | | CYP121 in complex with 3-bromo dicyclotyrosine | | Descriptor: | 3-bromo dicyclotyrosine, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
3QB7
 
 | | Interleukin-4 mutant RGA bound to cytokine receptor common gamma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Bates, D.L, Junttila, I.S, Creusot, R.J, Moraga, I, Lupardus, P, Fathman, C.G, Paul, W.E, Garcia, K.C. | | Deposit date: | 2011-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Redirecting cell-type specific cytokine responses with engineered interleukin-4 superkines.
Nat.Chem.Biol., 8, 2012
|
|
8F6P
 
 | | Rat Cardiac Sodium Channel with Ranolazine Bound | | Descriptor: | (R)-ranolazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lenaeus, M.J, Tonggu, L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis for Inhibition of the Cardiac Sodium Channel by the Atypical Antiarrhythmic Drug Ranolazine
To Be Published
|
|
3GNF
 
 | | P1 Crystal structure of the N-terminal R1-R7 of murine MVP | | Descriptor: | Major vault protein | | Authors: | Querol-Audi, J, Casanas, A, Uson, I, Luque, D, Caston, J.R, Fita, I, Verdaguer, N. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Embo J., 28, 2009
|
|
4CI3
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
3H1L
 
 | | Chicken cytochrome BC1 complex with ascochlorin bound at QO and QI sites | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 3-chloro-4,6-dihydroxy-2-methyl-5-{(2E,4E)-3-methyl-5-[(1R,2R,6R)-1,2,6-trimethyl-3-oxocyclohexyl]penta-2,4-dien-1-yl}benzaldehyde, CARDIOLIPIN, ... | | Authors: | Berry, E.A, Huang, L.S, Minagawa, N. | | Deposit date: | 2009-04-12 | | Release date: | 2010-01-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Ascochlorin is a novel, specific inhibitor of the mitochondrial cytochrome bc(1) complex.
Biochim.Biophys.Acta, 1797, 2010
|
|
8EKY
 
 | | Cryo-EM structure of the human PRDX4-ErP46 complex | | Descriptor: | Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
5X9R
 
 | | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS | | Descriptor: | CITRATE ANION, Citrate-sodium symporter, beta-D-glucopyranose | | Authors: | Jin, M.S, Kim, J.W, Kim, S, Kim, S, Lee, H, Lee, J.-O. | | Deposit date: | 2017-03-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS
Sci Rep, 7, 2017
|
|
6LKM
 
 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 5-chloro-N-((1S,2R)-2-(6-fluoro-2,3-dimethylphenyl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)-4-methyl-3,4-dihydro-2H-benzo[b][1,4]oxazine-8-sulfonamide | | Descriptor: | 5-chloro-N-((1S,2R)-2-(6-fluoro-2,3-dimethylphenyl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)-4-methyl-3,4-dihydro-2H-benzo[b][1,4]oxazine-8-sulfonamide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
3LIM
 
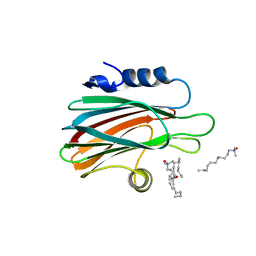 | | Crystal structure of the pore forming toxin frac from sea anemone actinia fragacea | | Descriptor: | Fragaceatoxin C, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Mechaly, A.E, Bellomio, A, Morante, K, Gonzalez-Manas, J.M, Guerin, D.M.A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins.
Structure, 19, 2011
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
3LJP
 
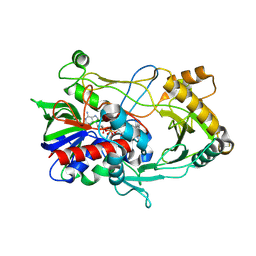 | | Crystal structure of choline oxidase V464A mutant | | Descriptor: | Choline oxidase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Finnegan, S, Agniswamy, J, Weber, I.T, Giovanni, G. | | Deposit date: | 2010-01-26 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of valine 464 in the flavin oxidation reaction catalyzed by choline oxidase.
Biochemistry, 49, 2010
|
|
6S0Q
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from 50,000 diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6LLX
 
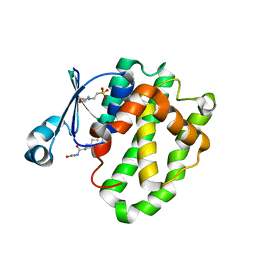 | |
4COB
 
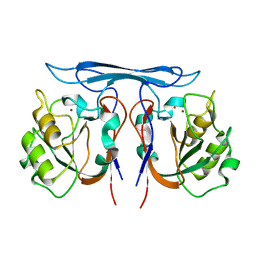 | | Crystal structure kynurenine formamidase from Pseudomonas aeruginosa | | Descriptor: | KYNURENINE FORMAMIDASE, ZINC ION | | Authors: | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
3QFC
 
 | | Crystal Structure of Human NADPH-Cytochrome P450 (V492E mutant) | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Marohnic, C, Panda, S.P, Masters, B.S, Kim, J.-J.P. | | Deposit date: | 2011-01-21 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8EOJ
 
 | | Microsomal triglyceride transfer protein | | Descriptor: | Microsomal triglyceride transfer protein large subunit, Protein disulfide-isomerase | | Authors: | Zhang, Z. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
6S12
 
 | | Erythromycin Resistant Staphylococcus aureus 50S ribosome (delta R88 A89 uL22). | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
6RVX
 
 | |
6LNG
 
 | | Rapid crystallization of streptavidin using charged peptides | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Minamihata, K, Tsukamoto, K, Adachi, M, Shimizu, R, Mishina, M, Kuroki, R, Nagamune, T. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8000015 Å) | | Cite: | Genetically fused charged peptides induce rapid crystallization of proteins.
Chem.Commun.(Camb.), 56, 2020
|
|
6RYF
 
 | | High-resolution crystal structure of ERAP1 in complex with 15mer phosphinic peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Mechanism for antigenic peptide selection by endoplasmic reticulum aminopeptidase 1.
Proc.Natl.Acad.Sci.USA, 2019
|
|
6S33
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with Protocatechuate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
8ET7
 
 | | Cryo-EM structure of the organic cation transporter 1 in complex with diphenhydramine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-[2-(BENZHYDRYLOXY)ETHYL]-N,N-DIMETHYLAMINE, OCT1 | | Authors: | Suo, Y, Wright, N.J, Lee, S.-Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Molecular basis of polyspecific drug and xenobiotic recognition by OCT1 and OCT2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
