1KHM
 
 | | C-TERMINAL KH DOMAIN OF HNRNP K (KH3) | | Descriptor: | PROTEIN (HNRNP K) | | Authors: | Baber, J, Libutti, D, Levens, D, Tjandra, N. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor.
J.Mol.Biol., 289, 1999
|
|
2W5H
 
 | | Human Nek2 kinase Apo | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Bayliss, R. | | Deposit date: | 2008-12-10 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Insights Into the Conformational Variability and Regulation of Human Nek2 Kinase.
J.Mol.Biol., 386, 2009
|
|
2ED9
 
 | |
1KMF
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Xu, B, Hua, Q.X, Nakagawa, S.H, Jia, W, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-09 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Chiral mutagenesis of insulin's hidden receptor-binding surface: structure of an allo-isoleucine(A2) analogue.
J.Mol.Biol., 316, 2002
|
|
2BAL
 
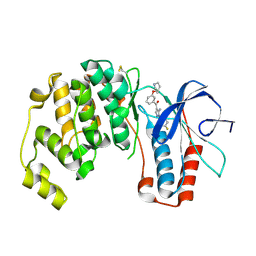 | | p38alpha MAP kinase bound to pyrazoloamine | | Descriptor: | Mitogen-activated protein kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL][3-(PIPERIDIN-4-YLOXY)PHENYL]METHANONE | | Authors: | Gerhardt, S, Pauptit, R.A, Read, J, Tucker, J, Norman, R.A, Breed, J. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase.
Biochemistry, 44, 2005
|
|
1YHV
 
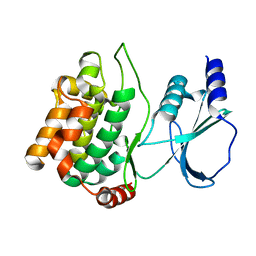 | |
2W5A
 
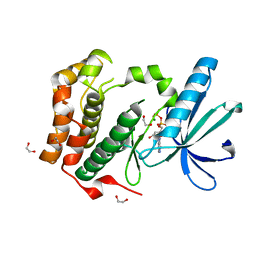 | | Human Nek2 kinase ADP-bound | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Westwood, I, Bayliss, R. | | Deposit date: | 2008-12-08 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Insights Into the Conformational Variability and Regulation of Human Nek2 Kinase.
J.Mol.Biol., 386, 2009
|
|
2B9J
 
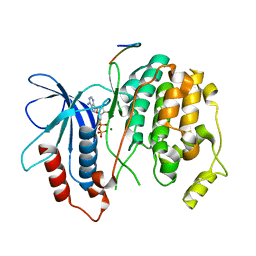 | | Crystal structure of Fus3 with a docking motif from Far1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cyclin-dependent kinase inhibitor FAR1, MAGNESIUM ION, ... | | Authors: | Remenyi, A, Good, M.C, Bhattacharyya, R.P, Lim, W.A. | | Deposit date: | 2005-10-11 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network.
Mol.Cell, 20, 2005
|
|
1YI3
 
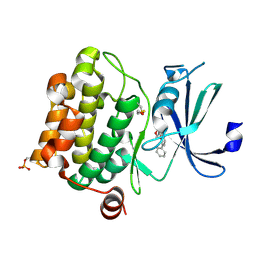 | | Crystal Structure of Pim-1 bound to LY294002 | | Descriptor: | 2-MORPHOLIN-4-YL-7-PHENYL-4H-CHROMEN-4-ONE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | Deposit date: | 2005-01-11 | | Release date: | 2005-01-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
1KJ7
 
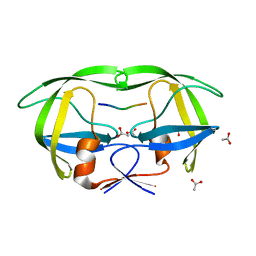 | |
1KMQ
 
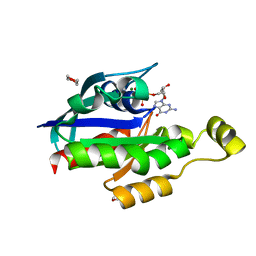 | | Crystal Structure of a Constitutively Activated RhoA Mutant (Q63L) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Longenecker, K, Read, P, Lin, S.-K, Somlyo, A.P, Nakamoto, R.K, Derewenda, Z.S. | | Deposit date: | 2001-12-17 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a constitutively activated RhoA mutant (Q63L) at 1.55 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3IPH
 
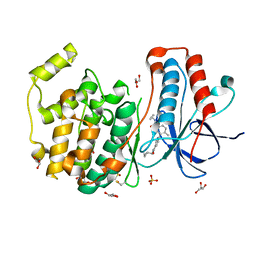 | | Crystal structure of p38 in complex with a biphenylamide inhibitor | | Descriptor: | 6-[5-(cyclopropylcarbamoyl)-2-methylphenyl]-N-(cyclopropylmethyl)pyridine-3-carboxamide, GLYCEROL, Mitogen-activated protein kinase 14, ... | | Authors: | Somers, D.O. | | Deposit date: | 2009-08-17 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | p38alpha mitogen-activated protein kinase inhibitors: optimization of a series of biphenylamides to give a molecule suitable for clinical progression.
J.Med.Chem., 52, 2009
|
|
1YZN
 
 | | GppNHp-Bound Ypt1p GTPase | | Descriptor: | GTP-binding protein YPT1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1KJF
 
 | |
1KJG
 
 | |
1Z3L
 
 | | X-Ray Crystal Structure of a Mutant Ribonuclease S (F8Anb) | | Descriptor: | Ribonuclease pancreatic, S-Peptide, S-Protein, ... | | Authors: | Das, M, Vasudeva Rao, B, Ghosh, S, Varadarajan, R. | | Deposit date: | 2005-03-14 | | Release date: | 2005-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Attempts to delineate the relative contributions of changes in hydrophobicity and packing to changes in stability of ribonuclease S mutants.
Biochemistry, 44, 2005
|
|
2WMR
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 5,6,7,8-TETRAHYDRO[1]BENZOTHIENO[2,3-D]PYRIMIDIN-4(3H)-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
2B53
 
 | |
2B0G
 
 | |
1KT6
 
 | | Crystal structure of bovine holo-RBP at pH 9.0 | | Descriptor: | RETINOL, plasma retinol-binding protein | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution Structures of Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
1Z8P
 
 | | Ferrous dioxygen complex of the A245S cytochrome P450eryF | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, 6-deoxyerythronolide B hydroxylase, OXYGEN MOLECULE, ... | | Authors: | Poulos, T.L, Nagano, S. | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the ferrous dioxygen complex of wild-type cytochrome P450eryF and its mutants, A245S and A245T: investigation of the proton transfer system in P450eryF.
J.Biol.Chem., 280, 2005
|
|
1Z5F
 
 | | Solution Structure of the Cytotoxic RC-RNase 3 with a Pyroglutamate Residue at the N-terminus | | Descriptor: | RC-RNase 3 | | Authors: | Lou, Y.C, Huang, Y.C, Pan, Y.R, Chen, C, Liao, Y.D. | | Deposit date: | 2005-03-18 | | Release date: | 2006-02-28 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Roles of N-terminal pyroglutamate in maintaining structural integrity and pKa values of catalytic histidine residues in bullfrog ribonuclease 3
J.Mol.Biol., 355, 2006
|
|
2B7F
 
 | | Crystal structure of human T-cell leukemia virus protease, a novel target for anti-cancer design | | Descriptor: | (ACE)APQV(STA)VMHP peptide, HTLV protease, PHOSPHATE ION | | Authors: | Li, M, Laco, G.S, Jaskolski, M, Rozycki, J, Alexandratos, J, Wlodawer, A, Gustchina, A. | | Deposit date: | 2005-10-04 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human T cell leukemia virus protease, a novel target for anticancer drug design
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B7Z
 
 | | Structure of HIV-1 protease mutant bound to indinavir | | Descriptor: | HIV-1 protease, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE | | Authors: | Clemente, J.C, Stow, L.R, Janka, L.K, Jeung, J.A, Desai, K.A, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2005-10-05 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vivo, kinetic, and structural analysis of the development of ritonavir resistance
To be Published
|
|
1Z6X
 
 | | Structure Of Human ADP-Ribosylation Factor 4 | | Descriptor: | ADP-ribosylation factor 4, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure Of Human ADP-Ribosylation Factor 4
To be Published
|
|
