6CZF
 
 | | The structure of E. coli PurF in complex with ppGpp-Mg | | Descriptor: | Amidophosphoribosyltransferase, GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, B, Grant, R.A, Laub, M.T. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Affinity-based capture and identification of protein effectors of the growth regulator ppGpp.
Nat. Chem. Biol., 15, 2019
|
|
8FV9
 
 | | E coli. CTP synthase in complex with F-dCTP | | Descriptor: | 2'-deoxy-2'-fluorocytidine 5'-(tetrahydrogen triphosphate), CTP synthase, MALONIC ACID | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FVB
 
 | | E coli. CTP synthase in complex with CTP | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MALONIC ACID | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FV6
 
 | | E coli. CTP synthase in complex with dF-dCTP | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), CTP synthase, MALONIC ACID, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FVE
 
 | | E coli. CTP synthase in complex with CTP (potassium malonate + 100 mM MgCl2) | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
8FV8
 
 | | E coli. CTP synthase in complex with dF-dCTP + ADP | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), ADENOSINE-5'-DIPHOSPHATE, CTP synthase, ... | | Authors: | Holyoak, T, McLeod, M.J, Tran, N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A metal-dependent conformational change provides a structural basis for the inhibition of CTP synthase by gemcitabine-5'-triphosphate.
Protein Sci., 32, 2023
|
|
2IU3
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase.
J. Biol. Chem., 282, 2007
|
|
2IU0
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
2O4C
 
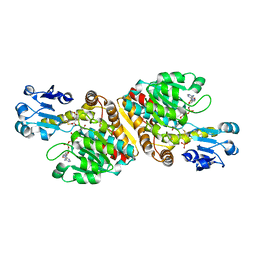 | | Crystal Structure of D-Erythronate-4-phosphate Dehydrogenase Complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ha, J.Y, Lee, J.H, Kim, K.H, Kim, D.J, Lee, H.H, Kim, H.K, Yoon, H.J, Suh, S.W. | | Deposit date: | 2006-12-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Erythronate-4-phosphate Dehydrogenase Complexed with NAD
J.Mol.Biol., 366, 2007
|
|
2NTM
 
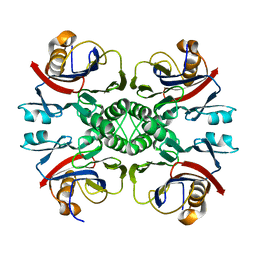 | |
2NTK
 
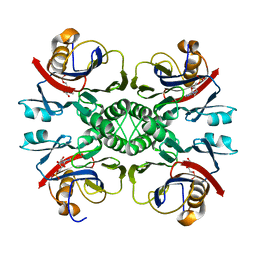 | | Crystal structure of PurO/IMP from Methanothermobacter thermoautotrophicus | | Descriptor: | IMP cyclohydrolase, INOSINIC ACID | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
4EW1
 
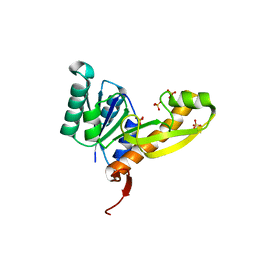 | | High resolution structure of human glycinamide ribonucleotide transformylase in apo form. | | Descriptor: | PHOSPHATE ION, SULFATE ION, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Connelly, S, DeMartino, K, Boger, D.L, Wilson, I.A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.522 Å) | | Cite: | Biological and Structural Evaluation of 10R- and 10S-Methylthio-DDACTHF Reveals a New Role for Sulfur in Inhibition of Glycinamide Ribonucleotide Transformylase.
Biochemistry, 52, 2013
|
|
4EW2
 
 | | The structure of human glycinamide ribonucleotide transformylase in complex with 10S-methylthio-DDATHF. | | Descriptor: | N-({4-[(1S)-4-(2,4-diamino-6-oxo-1,6-dihydropyrimidin-5-yl)-1-(methylsulfanyl)butyl]phenyl}carbonyl)-L-glutamic acid, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Connelly, S, DeMartino, K, Boger, D.L, Wilson, I.A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Biological and Structural Evaluation of 10R- and 10S-Methylthio-DDACTHF Reveals a New Role for Sulfur in Inhibition of Glycinamide Ribonucleotide Transformylase.
Biochemistry, 52, 2013
|
|
4FE2
 
 | | X-Ray Structure of SAICAR Synthetase (PurC) from Streptococcus pneumoniae complexed with AIR, ADP, Asp and Mg2+ | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wolf, N, Abad-Zapatero, C, Johnson, M.E, Fung, L.M.-W. | | Deposit date: | 2012-05-29 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structures of SAICAR synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg(2+), AIR and Asp.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6F4L
 
 | |
4EW3
 
 | | The structure of human glycinamide ribonucleotide transformylase in complex with 10R-methylthio-DDATHF. | | Descriptor: | N-({4-[(1R)-4-(2,4-diamino-6-oxo-1,6-dihydropyrimidin-5-yl)-1-(methylsulfanyl)butyl]phenyl}carbonyl)-L-glutamic acid, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Connelly, S, DeMartino, K, Boger, D.L, Wilson, I.A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Biological and Structural Evaluation of 10R- and 10S-Methylthio-DDACTHF Reveals a New Role for Sulfur in Inhibition of Glycinamide Ribonucleotide Transformylase.
Biochemistry, 52, 2013
|
|
8SBR
 
 | |
3ORS
 
 | | Crystal Structure of N5-Carboxyaminoimidazole Ribonucleotide Mutase from Staphylococcus aureus | | Descriptor: | N5-Carboxyaminoimidazole Ribonucleotide Mutase, SULFATE ION | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3ORQ
 
 | | Crystal Structure of N5-Carboxyaminoimidazole synthetase from Staphylococcus aureus complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4NYE
 
 | | Structures of SAICAR Synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg2+, AIR and L-Asp | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Fung, L.W.-M, Johnson, M.E, Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2013-12-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of SAICAR synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg(2+), AIR and Asp.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6VI8
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a superoxo bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VIA
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a seven-membered lactone bound structure | | Descriptor: | (2R,3E)-2-hydroxy-3-imino-2,3-dihydrooxepine-4-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3O8A
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with novel Inhibitor Genz667348 | | Descriptor: | Dihydroorotate dehydrogenase homolog, mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Booker, M.L, Phillips, M.A. | | Deposit date: | 2010-08-02 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase with anti-malarial activity in the mouse model.
J.Biol.Chem., 285, 2010
|
|
3QW3
 
 | | Structure of Leishmania donovani OMP decarboxylase | | Descriptor: | Orotidine-5-phosphate decarboxylase/orotate phosphoribosyltransferase, putative (Ompdcase-oprtase, putative), ... | | Authors: | French, J.B, Ealick, S.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization.
J.Biol.Chem., 286, 2011
|
|
6VI6
 
 | | Observing a ring-cleaving dioxygenase in action through a crystalline lens - a substrate monodentately bound structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Wang, Y, Liu, F, Yang, Y, Liu, A. | | Deposit date: | 2020-01-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Observing 3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
