5DGI
 
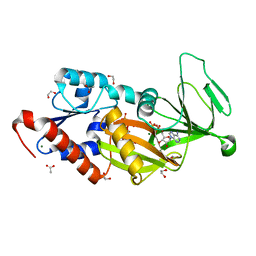 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 3,5-(PCP)2-IP4 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-di-methylenebisphosphonate inositol tetrakisphosphate, ACETATE ION, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cellular Cations Control Conformational Switching of Inositol Pyrophosphate Analogues.
Chemistry, 22, 2016
|
|
7F9W
 
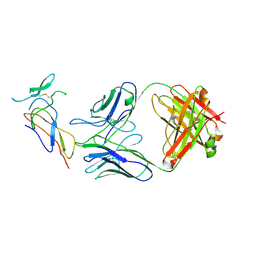 | | CD25 in complex with Fab | | Descriptor: | Heavy chain of Fab, Interleukin-2 receptor subunit alpha, Light chain of Fab | | Authors: | Liu, C. | | Deposit date: | 2021-07-05 | | Release date: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Two novel human anti-CD25 antibodies with antitumor activity inversely related to their affinity and in vitro activity.
Sci Rep, 11, 2021
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7TU6
 
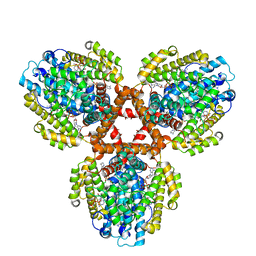 | | Structure of the L. blandensis dGTPase bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU7
 
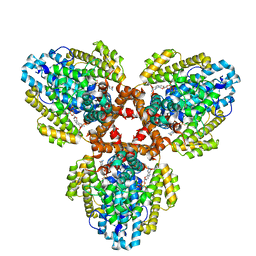 | | Structure of the L. blandensis dGTPase H125A mutant bound to dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU8
 
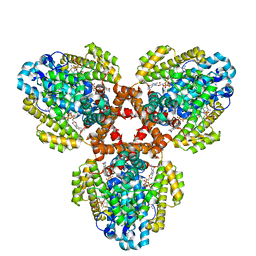 | | Structure of the L. blandensis dGTPase H125A mutant bound to dGTP and dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU5
 
 | | Structure of the L. blandensis dGTPase in the apo form | | Descriptor: | MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7CJR
 
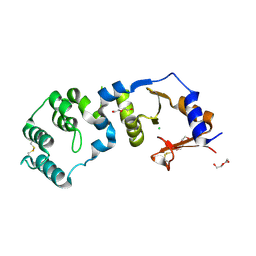 | | Crystal structure of a periplasmic sensor domain of histidine kinase VbrK | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Histidine kinase | | Authors: | Goh, B.C, Chua, Y.K, Qian, X, Savko, M, Lescar, J. | | Deposit date: | 2020-07-12 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the periplasmic sensor domain of histidine kinase VbrK suggests indirect sensing of beta-lactam antibiotics.
J.Struct.Biol., 212, 2020
|
|
7F2H
 
 | |
7F2G
 
 | |
1MVT
 
 | | Analysis of Two Polymorphic Forms of a Pyrido[2,3-d]pyrimidine N9-C10 Reverse-Bridge Antifolate Binary Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',4',5'-TRIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate Reductase, SULFATE ION | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W.A, Gangjee, A. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of two polymorphic forms of a pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolate binary complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MVS
 
 | | Analysis of Two Polymorphic Forms of a Pyrido[2,3-d]pyrimidine N9-C10 Reverse-Bridge Antifolate Binary Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',4',5'-TRIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate Reductase, SULFATE ION | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W.A, Gangjee, A. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of two polymorphic forms of a pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolate binary complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2RDA
 
 | |
1PD9
 
 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-5-METHYL-6-[(3,4,5-TRIMETHOXY-N-METHYLANILINO)METHYL]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PD8
 
 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine Antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-5-METHYL-6-[(3,4,5-TRIMETHOXY-N-METHYLANILINO)METHYL]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PDB
 
 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine Antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | Dihydrofolate reductase | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2V9A
 
 | | Structure of Citrate-free Periplasmic Domain of Sensor Histidine Kinase CitA | | Descriptor: | SENSOR KINASE CITA | | Authors: | Sevvana, M, Vijayan, V, Zweckstetter, M, Reinelt, S, Madden, D.R, Sheldrick, G.M, Bott, M, Griesinger, C, Becker, S. | | Deposit date: | 2007-08-23 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Ligand-Induced Switch in the Periplasmic Domain of Sensor Histidine Kinase Cita.
J.Mol.Biol., 377, 2008
|
|
3BPP
 
 | |
2Z9I
 
 | |
3DBA
 
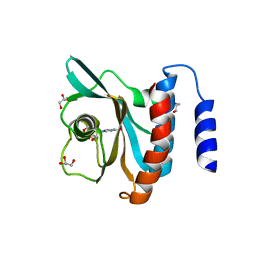 | | Crystal structure of the cGMP-bound GAF a domain from the photoreceptor phosphodiesterase 6C | | Descriptor: | Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha', GLYCEROL, GUANOSINE-3',5'-MONOPHOSPHATE | | Authors: | Martinez, S.E, Heikaus, C.C, Klevit, R.E, Beavo, J.A. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The Structure of the GAF A Domain from Phosphodiesterase 6C Reveals Determinants of cGMP Binding, a Conserved Binding Surface, and a Large cGMP-dependent Conformational Change.
J.Biol.Chem., 283, 2008
|
|
1DPT
 
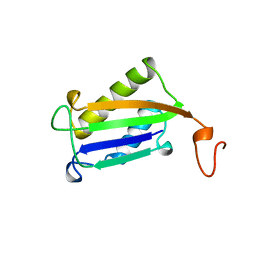 | | D-DOPACHROME TAUTOMERASE | | Descriptor: | D-DOPACHROME TAUTOMERASE | | Authors: | Sugimoto, H, Taniguchi, M, Nakagawa, A, Tanaka, I. | | Deposit date: | 1998-05-11 | | Release date: | 1999-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of human D-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 A resolution.
Biochemistry, 38, 1999
|
|
8ZWJ
 
 | |
8ZWK
 
 | |
9CK3
 
 | | Cryo-EM structure of in-vitro alpha-synuclein fibril | | Descriptor: | Alpha-synuclein | | Authors: | Sanchez, J.C, Borcik, C.G, Tonelli, M, Sibert, B, Rienstra, C.M, Wright, E.R. | | Deposit date: | 2024-07-08 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Multimodal High-Resolution Structure Determination Of Alpha-Synuclein Fibrils
To Be Published
|
|
9IJP
 
 | |
