5FKW
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | Descriptor: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
6N2T
 
 | |
5FKV
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.04 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
1HWT
 
 | | STRUCTURE OF A HAP1/DNA COMPLEX REVEALS DRAMATICALLY ASYMMETRIC DNA BINDING BY A HOMODIMERIC PROTEIN | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*C)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a HAP1-DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein.
Nat.Struct.Biol., 6, 1999
|
|
2HAP
 
 | | STRUCTURE OF A HAP1-18/DNA COMPLEX REVEALS THAT PROTEIN/DNA INTERACTIONS CAN HAVE DIRECT ALLOSTERIC EFFECTS ON TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of HAP1-18-DNA implicates direct allosteric effect of protein-DNA interactions on transcriptional activation.
Nat.Struct.Biol., 6, 1999
|
|
6LXN
 
 | |
4R65
 
 | | Ternary complex crystal structure of R258A mutant of DNA polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-induced DNA Polymerase beta Activation.
J.Biol.Chem., 289, 2014
|
|
4R66
 
 | | Ternary complex crystal structure of E295K mutant of DNA polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate-induced DNA Polymerase beta Activation.
J.Biol.Chem., 289, 2014
|
|
6MR7
 
 | | DNA polymerase beta substrate complex with templating adenine and incoming Fapy-dGTP analog | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-amino-5-(formylamino)-6-oxo-1,6-dihydropyrimidin-4-yl]-2,5-anhydro-1,3-dideoxy-6-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-ribo-hexitol, CALCIUM ION, ... | | Authors: | Freudenthal, B.D, Smith, M.R, Wilson, S.H, Beard, W.A. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | A guardian residue hinders insertion of a Fapy•dGTP analog by modulating the open-closed DNA polymerase transition.
Nucleic Acids Res., 47, 2019
|
|
4TNT
 
 | | Structure of the human mineralocorticoid receptor in complex with DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), Mineralocorticoid receptor, ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2014-06-04 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3938 Å) | | Cite: | Crystal structure of the mineralocorticoid receptor DNA binding domain in complex with DNA.
Plos One, 9, 2014
|
|
1EOO
 
 | | ECORV BOUND TO COGNATE DNA | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*AP*TP*AP*TP*CP*TP*TP*C)-3'), TYPE II RESTRICTION ENZYME ECORV | | Authors: | Horton, N.C, Perona, J.J. | | Deposit date: | 2000-03-23 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystallographic snapshots along a protein-induced DNA-bending pathway.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EOP
 
 | | ECORV BOUND TO COGNATE DNA | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*AP*TP*AP*TP*CP*TP*TP*C)-3'), TYPE II RESTRICTION ENZYME ECORV | | Authors: | Horton, N.C, Perona, J.J. | | Deposit date: | 2000-03-23 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic snapshots along a protein-induced DNA-bending pathway.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8V5O
 
 | | Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.99 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5N
 
 | | Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.56 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5M
 
 | | Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (9.22 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1E3M
 
 | | The crystal structure of E. coli MutS binding to DNA with a G:T mismatch | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP* GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP* TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Perrakis, A, Enzlin, J.H, Winterwerp, H.H.K, De Wind, N, Sixma, T.K. | | Deposit date: | 2000-06-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of DNA Mismatch Repair Protein Muts Binding to a G X T Mismatch
Nature, 407, 2000
|
|
6O3T
 
 | | Structural basis of FOXC2 and DNA interactions | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein C2 | | Authors: | Nam, H.-J, Li, S. | | Deposit date: | 2019-02-27 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Crystal Structure of FOXC2 in Complex with DNA Target.
Acs Omega, 4, 2019
|
|
1IF1
 
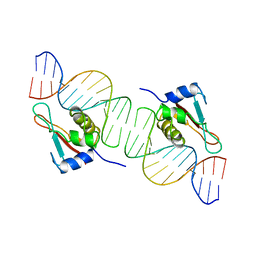 | | INTERFERON REGULATORY FACTOR 1 (IRF-1) COMPLEX WITH DNA | | Descriptor: | DNA (26-MER), PROTEIN (INTERFERON REGULATORY FACTOR 1) | | Authors: | Escalante, C.R, Yie, J, Thanos, D, Aggarwal, A. | | Deposit date: | 1997-09-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of IRF-1 with bound DNA reveals determinants of interferon regulation.
Nature, 391, 1998
|
|
3CO7
 
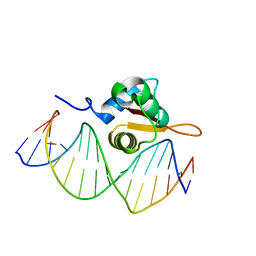 | | Crystal Structure of FoxO1 DBD Bound to DBE2 DNA | | Descriptor: | DNA (5'-D(*DCP*DAP*DAP*DAP*DAP*DTP*DGP*DTP*DAP*DAP*DAP*DCP*DAP*DAP*DGP*DA)-3'), DNA (5'-D(*DTP*DCP*DTP*DTP*DGP*DTP*DTP*DTP*DAP*DCP*DAP*DTP*DTP*DTP*DTP*DG)-3'), Forkhead box protein O1 | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
4XIU
 
 | |
8SY7
 
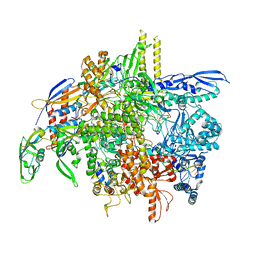 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-STP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
1PYI
 
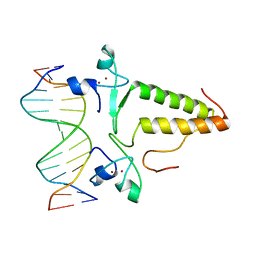 | |
1CMA
 
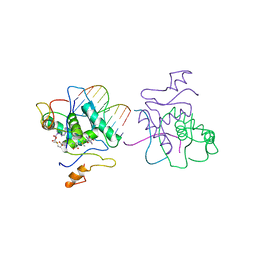 | | MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE | | Descriptor: | DNA (5'-D(*AP*GP*AP*CP*GP*TP*CP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*AP*CP*GP*TP*CP*T)-3'), PROTEIN (MET REPRESSOR), ... | | Authors: | Somers, W.S, Phillips, S.E.V. | | Deposit date: | 1992-08-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands.
Nature, 359, 1992
|
|
1HF0
 
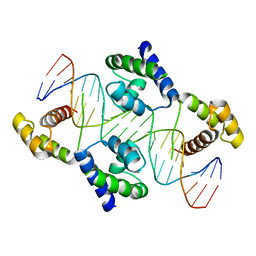 | | Crystal structure of the DNA-binding domain of Oct-1 bound to DNA as a dimer | | Descriptor: | DNA 5'-D(*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP* CP*AP*AP*AP*TP*GP*GP*AP*G)-3', DNA 5'-D(*CP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP* TP*CP*AP*AP*AP*TP*GP*TP*G)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | Authors: | Remenyi, A, Tomilin, A, Pohl, E, Scholer, H.R, Wilmanns, M. | | Deposit date: | 2000-11-27 | | Release date: | 2001-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
8G9L
 
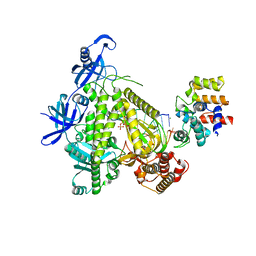 | | DNA initiation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
