6G73
 
 | | The dynamic nature of the VDAC1 channels in bilayers: human VDAC1 at 3.3 Angstrom resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Razeto, A, Gribbon, P, Loew, C. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | The dynamic nature of the VDAC1 channels in bilayers as revealed by two crystal structures of the human isoform in bicelles at 2.7 and 3.3 Angstrom resolution: implications for VDAC1 voltage-dependent mechanism and for its oligomerization
To Be Published
|
|
6G7H
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: resting state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7O
 
 | | Crystal structure of human alkaline ceramidase 3 (ACER3) at 2.7 Angstrom resolution | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Alkaline ceramidase 3,Soluble cytochrome b562, CALCIUM ION, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Healey, R.D, Sounier, R, Grison, C, Hoh, F, Basu, S, Granier, S. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a human intramembrane ceramidase explains enzymatic dysfunction found in leukodystrophy.
Nat Commun, 9, 2018
|
|
6GBZ
 
 | | 50S ribosomal subunit assembly intermediate state 5 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
6GA5
 
 | | Bacteriorhodopsin, 3 ps state, REAL-SPACE REFINEMED AGAINST 10% EXTRAPOLATED MAP | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAF
 
 | | BACTERIORHODOPSIN, 590 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GXN
 
 | | Cryo-EM structure of an E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and Pint-tRNA (State III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
5D80
 
 | | Crystal Structure of Yeast V1-ATPase in the Autoinhibited Form | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit D, ... | | Authors: | Oot, R.A, Kane, P.M, Berry, E.A, Wilkens, S. | | Deposit date: | 2015-08-14 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (6.202 Å) | | Cite: | Crystal structure of yeast V1-ATPase in the autoinhibited state.
Embo J., 35, 2016
|
|
5DG1
 
 | | Sugar binding protein - human galectin-2 | | Descriptor: | Galectin-2, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human galectin-2 interacts with carbohydrates and peptides non-classically: new insight from X-ray crystallography and hemagglutination.
Acta Biochim.Biophys.Sin., 2016
|
|
5D3G
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a Novel 38-mer Hairpin Template-Primer DNA Aptamer | | Descriptor: | DNA aptamer (38-MER), GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer.
Protein Sci., 25, 2016
|
|
6GJU
 
 | | human NBD1 of CFTR in complex with nanobodies T2a and T4 | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
5CQB
 
 | | Crystal structure of E. coli undecaprenyl pyrophosphate synthase | | Descriptor: | Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific), TRIETHYLENE GLYCOL | | Authors: | Worrall, L.J, Conrady, D.G, Strynadka, N.C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6GQZ
 
 | | Petrobactin-binding engineered lipocalin without ligand | | Descriptor: | Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A. | | Deposit date: | 2018-06-08 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reprogramming Human Siderocalin To Neutralize Petrobactin, the Essential Iron Scavenger of Anthrax Bacillus.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GS7
 
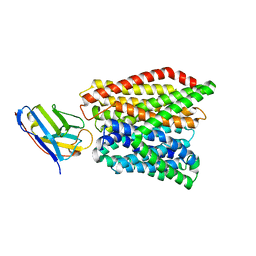 | | Crystal structure of peptide transporter DtpA-nanobody in glycine buffer | | Descriptor: | Dipeptide and tripeptide permease A, nanobody | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
5CYQ
 
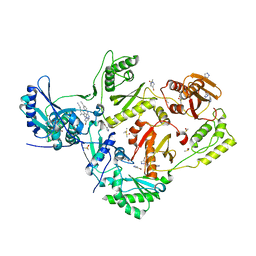 | | HIV-1 reverse transcriptase complexed with 4-bromopyrazole | | Descriptor: | 4-bromo-1H-pyrazole, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, BROMIDE ION, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Rapid experimental SAD phasing and hot-spot identification with halogenated fragments.
IUCrJ, 3, 2016
|
|
6GTA
 
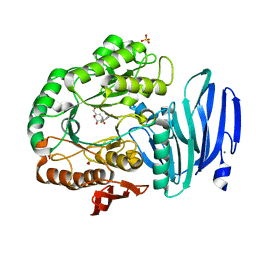 | | Alpha-galactosidase mutant D378A from Thermotoga maritima in complex with intact cyclohexene-based carbasugar mimic of galactose with 3,5 difluorophenyl leaving group | | Descriptor: | (1~{R},2~{S},3~{S},6~{S})-6-[3,5-bis(fluoranyl)phenoxy]-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Pengelly, R.J. | | Deposit date: | 2018-06-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
5CXY
 
 | | Structure of a Glycosyltransferase in Complex with Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Volkers, G, Strynadka, N.C.J. | | Deposit date: | 2015-07-29 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | To be published.
To be published
|
|
6GJS
 
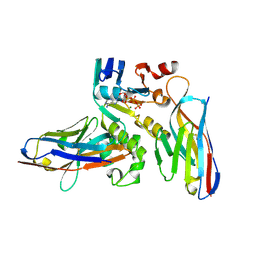 | | Human NBD1 of CFTR in complex with nanobodies D12 and T4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6GK4
 
 | | Human NBD1 of CFTR in complex with nanobodies D12 and T8 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-18 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6GS1
 
 | | Crystal structure of peptide transporter DtpA-nanobody in MES buffer | | Descriptor: | Dipeptide and tripeptide permease A, Nanobody 00 | | Authors: | Ural-Blimke, Y, Flayhan, A, Loew, C, Quistgaard, E.M. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
6GT3
 
 | | Crystal Structure of the A2A-StaR2-bRIL562 in complex with AZD4635 at 2.0A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2-chloranyl-6-methyl-pyridin-4-yl)-5-(4-fluorophenyl)-1,2,4-triazin-3-amine, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Borodovsky, A, Wang, Y, Deng, N, Ye, M, Stephen, T.L, Goodwin, K, Goodwin, R, Strittmatter, N, Shaw, J, Sachsenmeier, K, Clarke, J.D, Hay, C, Reimer, C, Andrews, S.P, Brown, G.A, Congreve, M, Cheng, R.K.Y, Dore, A.S, Mason, J.S, Marshall, F.H, Weir, M.P, Lyne, P, Woessner, R. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small molecule AZD4635 inhibitor of A2AR signaling rescues immune cell function including CD103+ dendritic cells enhancing anti-tumor immunity
J Immunother Cancer, 2020
|
|
5CU5
 
 | | Crystal structure of ERAP2 without catalytic Zn(II) atom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Mathioudakis, N, Giastas, P, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (ER) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
5CYM
 
 | | HIV-1 reverse transcriptase complexed with 4-iodopyrazole | | Descriptor: | 1,2-ETHANEDIOL, 4-IODOPYRAZOLE, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-30 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rapid experimental SAD phasing and hot spot identification with halogenated fragments
Iucrj, 3, 2016
|
|
5CRX
 
 | |
