6FBF
 
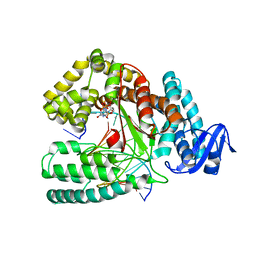 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the fourth primer nucleotide. | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*CP*GP*GP*TP*GP*CP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*GP*CP*AP*(OH3)P*CP*GP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7SAA
 
 | |
7SL5
 
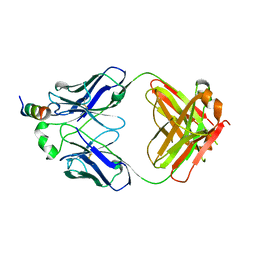 | |
6FB2
 
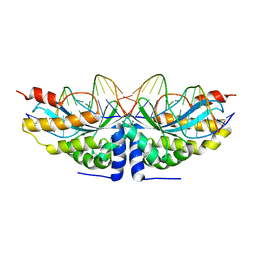 | |
6FBC
 
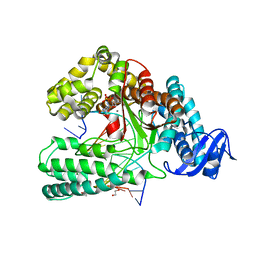 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification at the 3'-terminus of the primer. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
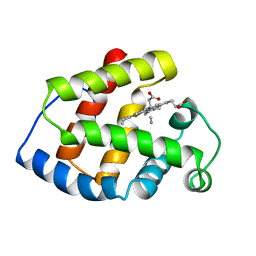
2LHB
 
 | |
4XJR
 
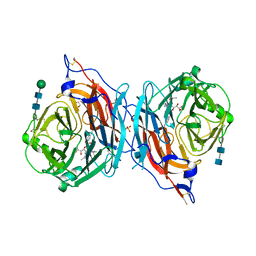 | | The catalytic mechanism of human parainfluenza virus type 3 haemagglutinin-neuraminidase revealed | | Descriptor: | (6R)-2,6-anhydro-3,4,5-trideoxy-6-[(2S)-2,3-dihydroxypropanoyl]-3-fluoro-5-[(2-methylpropanoyl)amino]-4-triaza-1,2-dien -2-ium-1-yl-L-gulonic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, El-Deeb, I, Guillon, P, Carroux, C, Chavas, L, von Itzstein, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The catalytic mechanism of human parainfluenza virus type 3 haemagglutinin-neuraminidase revealed.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3RB5
 
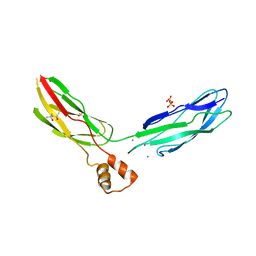 | | Crystal structure of calcium binding domain CBD12 of CALX1.1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Na/Ca exchange protein, ... | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2011-03-28 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of the Ca(2+) Inhibitory Mechanism of Drosophila Na(+)/Ca(2+) Exchanger CALX and Its Modification by Alternative Splicing.
Structure, 19, 2011
|
|
4LE4
 
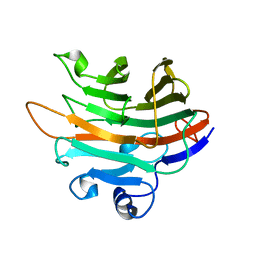 | | Crystal structure of PaGluc131A with cellotriose | | Descriptor: | Beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, T, Chan, H.C, Huang, C.H, Ko, T.P, Huang, T.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a GH131 beta-Glucanase Catalytic Domain from Podospora anserina in Complex with Cellotriose
To be Published
|
|
4LO5
 
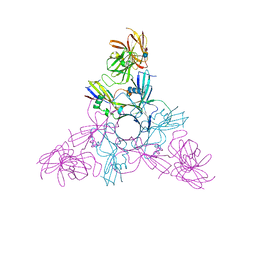 | | HA70-alpha2,3-SiaLC | | Descriptor: | CHLORIDE ION, HA-70, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
3KBM
 
 | |
3KCL
 
 | |
3R29
 
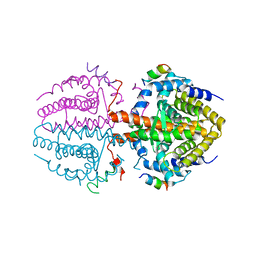 | | Crystal structure of RXRalpha ligand-binding domain complexed with corepressor SMRT2 | | Descriptor: | Nuclear receptor corepressor 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Chen, L, Chen, J, Jiang, H, Shen, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for retinoic x receptor repression on the tetramer.
J.Biol.Chem., 286, 2011
|
|
3R2A
 
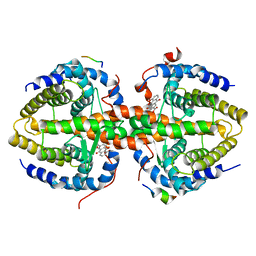 | | Crystal structure of RXRalpha ligand-binding domain complexed with corepressor SMRT2 and antagonist rhein | | Descriptor: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Nuclear receptor corepressor 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Chen, L, Chen, J, Jiang, H, Shen, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for retinoic x receptor repression on the tetramer.
J.Biol.Chem., 286, 2011
|
|
3RTI
 
 | |
4LO2
 
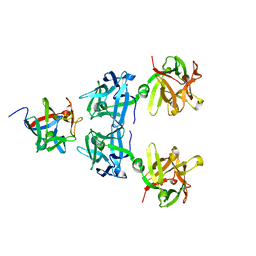 | | HA17-HA33-Lac | | Descriptor: | HA-17, HA-33, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
3RLF
 
 | |
3RO4
 
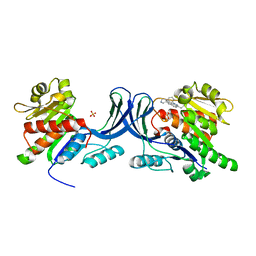 | |
3RT6
 
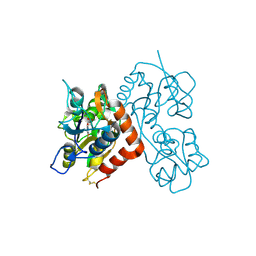 | |
3RGZ
 
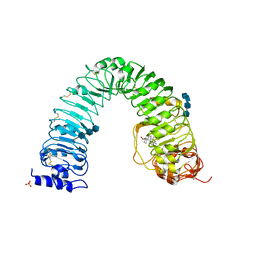 | | Structural insight into brassinosteroid perception by BRI1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chai, J, Han, Z, She, J, Wang, J, Cheng, W. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural insight into brassinosteroid perception by BRI1.
Nature, 474, 2011
|
|
3KLY
 
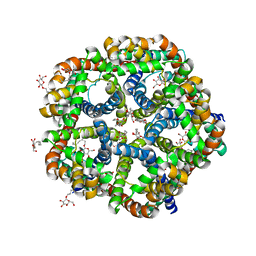 | |
3RT8
 
 | |
3L38
 
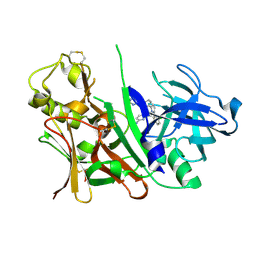 | | Bace1 in complex with the aminopyridine Compound 44 | | Descriptor: | 6-({2-(2-chlorophenyl)-5-[4-(pyrimidin-5-yloxy)phenyl]-1H-pyrrol-1-yl}methyl)pyridin-2-amine, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel pyrrolyl 2-aminopyridines as potent and selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7SIM
 
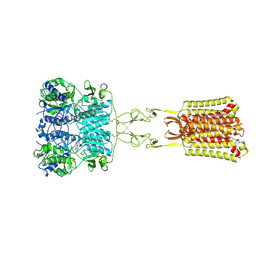 | | Structure of positive allosteric modulator-free active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3KNQ
 
 | |
