4MJH
 
 | |
4MK8
 
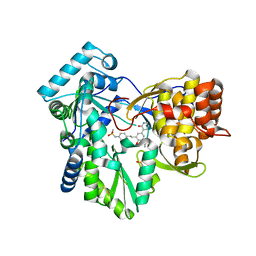 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with inhibitor 4 (N-(4-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]ethyl}phenyl)methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-(4-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]ethyl}phenyl)methanesulfonamide, ... | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2013-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of a Novel Series of Potent Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B.
J.Med.Chem., 56, 2013
|
|
4MOV
 
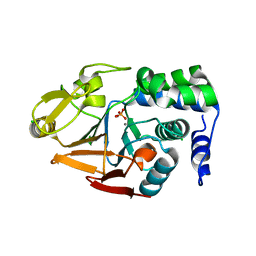 | | 1.45 A Resolution Crystal Structure of Protein Phosphatase 1 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4503 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MP4
 
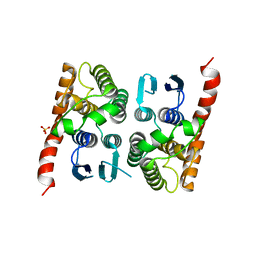 | | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure
To be Published
|
|
4MP8
 
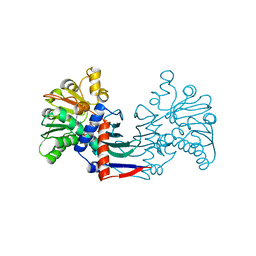 | | Staphyloferrin B precursor biosynthetic enzyme SbnB bound to malonate and NAD+ | | Descriptor: | MALONATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative ornithine cyclodeaminase | | Authors: | Grigg, J.C, Kobylarz, M.J, Rai, D.K, Murphy, M.E.P. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of L-2,3-diaminopropionic Acid, a siderophore and antibiotic precursor.
Chem.Biol., 21, 2014
|
|
4MQG
 
 | |
4MRM
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist phaclofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MT5
 
 | | Crystal structure of Mub-RV | | Descriptor: | Mucus binding proteinn | | Authors: | Etzold, S, Juge, N, Hemmings, A.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for adaptation of lactobacilli to gastrointestinal mucus.
ENVIRON.MICROBIOL., 16, 2014
|
|
4MU6
 
 | |
4MK3
 
 | | Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h) | | Descriptor: | Glutathione S-transferase, L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE, MAGNESIUM ION, ... | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h)
To be published
|
|
4MKB
 
 | |
4MX5
 
 | | Structure of ricin A chain bound with benzyl-(2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)ethyl)carbamate | | Descriptor: | Ricin A chain, benzyl (2-{[(2-amino-4-oxo-3,4-dihydropteridin-7-yl)carbonyl]amino}ethyl)carbamate | | Authors: | Jasheway, K.R, Robertus, J.D, Wiget, P.A, Manzano, L.A, Pruet, J.M, Gao, G, Saito, R, Monzingo, A.F, Anslyn, E.V. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Sulfur incorporation generally improves Ricin inhibition in pterin-appended glycine-phenylalanine dipeptide mimics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4MNC
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P21 | | Descriptor: | BENZOYL-FORMIC ACID, SULFATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MJQ
 
 | | E. coli sliding clamp in complex with Bromfenac | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs.
Chem.Biol., 21, 2014
|
|
4MK9
 
 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with inhibitor 12 (N-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1,3-benzoxazol-5-yl}methanesulfonamide) | | Descriptor: | GLYCEROL, N-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1,3-benzoxazol-5-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2013-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Novel Series of Potent Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B.
J.Med.Chem., 56, 2013
|
|
4MIB
 
 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with Compound 48 (N-({(3S)-1-[6-tert-butyl-5-methoxy-8-(2-oxo-1,2-dihydropyridin-3-yl)quinolin-3-yl]pyrrolidin-3-yl}methyl)methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, N-({(3S)-1-[6-tert-butyl-5-methoxy-8-(2-oxo-1,2-dihydropyridin-3-yl)quinolin-3-yl]pyrrolidin-3-yl}methyl)methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Harris, S.F, Villasenor, A.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of N-[4-[6-tert-butyl-5-methoxy-8-(6-methoxy-2-oxo-1H-pyridin-3-yl)-3-quinolyl]phenyl]methanesulfonamide (RG7109), a potent inhibitor of the hepatitis C virus NS5B polymerase.
J.Med.Chem., 57, 2014
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | Deposit date: | 2005-08-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
4V4J
 
 | | Interactions and Dynamics of the Shine-Dalgarno Helix in the 70S Ribosome. | | Descriptor: | 16S RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Trakhanov, S, Asahara, H, Laurberg, M, Noller, H.F. | | Deposit date: | 2007-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4V57
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with spectinomycin and neomycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Shoji, S, Holton, J.M, Fredrick, K, Cate, J.H.D. | | Deposit date: | 2007-07-21 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A steric block in translation caused by the antibiotic spectinomycin.
Acs Chem.Biol., 2, 2007
|
|
4V9H
 
 | | Crystal structure of the ribosome bound to elongation factor G in the guanosine triphosphatase state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tourigny, D.S, Fernandez, I.S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2013-03-25 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (2.857 Å) | | Cite: | Elongation factor G bound to the ribosome in an intermediate state of translocation.
Science, 340, 2013
|
|
4V6G
 
 | | Initiation complex of 70S ribosome with two tRNAs and mRNA. | | Descriptor: | 16S RRNA (E.COLI NUMBERING), 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jenner, L.B, Yusupova, G, Yusupov, M. | | Deposit date: | 2009-07-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural aspects of messenger RNA reading frame maintenance by the ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4V50
 
 | | Crystal Structure of Ribosome with messenger RNA and the Anticodon stem-loop of P-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Berk, V, Zhang, W, Pai, R.D, Cate, J.H.D. | | Deposit date: | 2006-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for mRNA and tRNA positioning on the ribosome.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4W90
 
 | | Crystal structure of Bacillus subtilis cyclic-di-AMP riboswitch ydaO | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.118 Å) | | Cite: | Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA.
Embo J., 33, 2014
|
|
4V94
 
 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
4V4I
 
 | | Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements. | | Descriptor: | 16S SMALL SUBUNIT RIBOSOMAL RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Trakhanov, S, Laurberg, M, Noller, H.F. | | Deposit date: | 2007-02-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements
Cell(Cambridge,Mass.), 126, 2006
|
|
