4BWL
 
 | | Structure of the Y137A mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate, N-acetyl-D-mannosamine and N- acetylneuraminic acid | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, 5-(acetylamino)-3,5-dideoxy-D-glycero-D-galacto-non-2-ulosonic acid, N-ACETYLNEURAMINATE LYASE, ... | | Authors: | Campeotto, I, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2013-07-03 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Reaction Mechanism of N-Acetylneuraminic Acid Lyase Revealed by a Combination of Crystallography, Qm/Mm Simulation and Mutagenesis.
Acs Chem.Biol., 9, 2014
|
|
6OF1
 
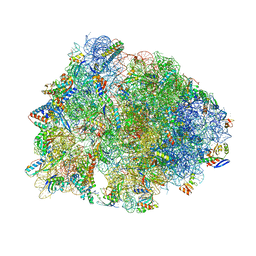 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with dirithromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khabibullina, N.F, Tereshchenkov, A.G, Komarova, E.S, Syroegin, E.A, Shiriaev, D.I, Paleskava, A, Kartsev, V.G, Bogdanov, A.A, Konevega, A.L, Dontsova, O.A, Sergiev, P.V, Osterman, I.A, Polikanov, Y.S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-04-17 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Dirithromycin Bound to the Bacterial Ribosome Suggests New Ways for Rational Improvement of Macrolides.
Antimicrob.Agents Chemother., 63, 2019
|
|
9B0T
 
 | | Cryo-EM structure of E227Q variant of uMtCK1 in complex with transition state analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-12 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
9B14
 
 | | Cryo-EM structure of human uMtCK1 in complex with transition state analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-13 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
9B0U
 
 | | Cryo-EM structure of E227Q variant of uMtCK1 incubated with ADP and phosphocreatine at pH 8.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-12 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
6O5S
 
 | |
5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
7NHN
 
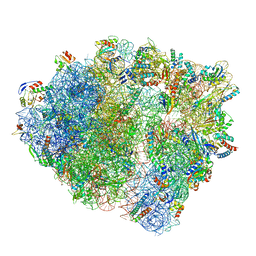 | | VgaL, an antibiotic resistance ABCF, in complex with 70S ribosome from Listeria monocytogenes | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Turnbull, K.J, Hauryliuk, V, Wilson, D.N. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
2C0B
 
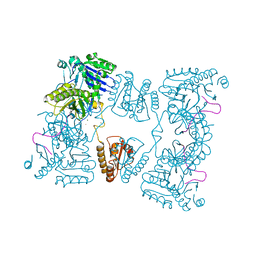 | | Catalytic domain of E. coli RNase E in complex with 13-mer RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP*UP*G)-3', MAGNESIUM ION, RIBONUCLEASE E, ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-08-30 | | Release date: | 2005-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2C4R
 
 | | Catalytic domain of E. coli RNase E | | Descriptor: | MAGNESIUM ION, RIBONUCLEASE E, SSRNA MOLECULE: 5'-R(*AP*CP*AP*GP*UP*AP*UP*UP*UP*GP)-3', ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Luisi, B.F. | | Deposit date: | 2005-10-21 | | Release date: | 2005-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2BX2
 
 | | Catalytic domain of E. coli RNase E | | Descriptor: | MAGNESIUM ION, RIBONUCLEASE E, RNA (5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP* UP*GP*UP*U)-3'), ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
3J2P
 
 | | CryoEM structure of Dengue virus envelope protein heterotetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Zhang, X, Ge, P, Yu, X, Brannan, J.M, Bi, G, Zhang, Q, Schein, S, Zhou, Z.H. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the mature dengue virus at 3.5-A resolution.
Nat.Struct.Mol.Biol., 20, 2012
|
|
7OVW
 
 | | Binding domain of botulinum neurotoxin E in complex with GD1a | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin type E | | Authors: | Masuyer, G, Stenmark, P. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Ganglioside Receptor Recognition by Botulinum Neurotoxin Serotype E.
Int J Mol Sci, 22, 2021
|
|
7NPK
 
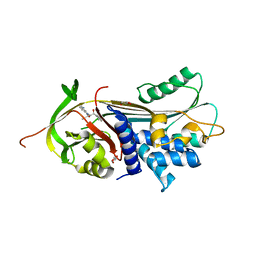 | | ALPHA-1 ANTITRYPSIN C232S COMPLEXED WITH CMPD3 | | Descriptor: | Alpha-1-antitrypsin, GLYCEROL, N-((1S,2R)-1-hydroxy-1-(o-tolyl)pentan-2-yl)-2-oxo-2,3-dihydrobenzo[d]oxazole-5-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The development of highly potent and selective small molecule correctors of Z alpha 1 -antitrypsin misfolding.
Bioorg.Med.Chem.Lett., 41, 2021
|
|
7RQ8
 
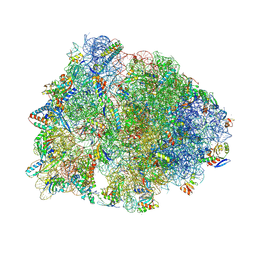 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
9BIB
 
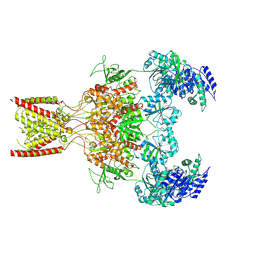 | | Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation, C1 symmetry | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2024-04-23 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Molecular mechanism of ligand gating and opening of NMDA receptor.
Nature, 632, 2024
|
|
7RQ9
 
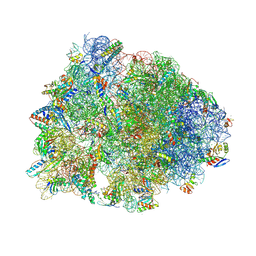 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
6ZKY
 
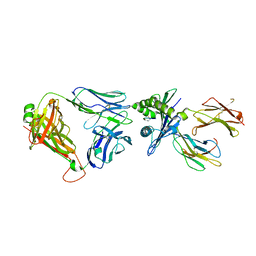 | | Crystal structure of InhA:01 TCR in complex with HLA-E (S147C) bound to InhA (53-61 H3C) | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], HLA class I histocompatibility antigen, ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
6ZKW
 
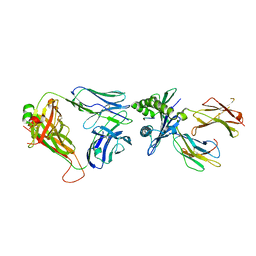 | | Crystal structure of InhA:01 TCR in complex with HLA-E bound to InhA (53-61) | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], HLA class I histocompatibility antigen, ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
6ZKX
 
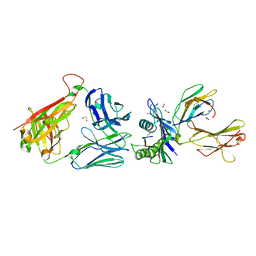 | | Crystal structure of InhA:01 TCR in complex with HLA-E (Y84C) bound to InhA (53-61 GCG) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
6ZKZ
 
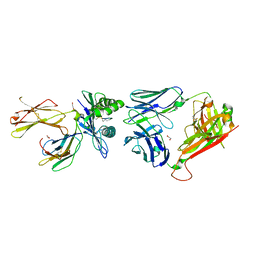 | | Crystal structure of InhA:01 TCR in complex with HLA-E (F116C) bound to InhA (53-61 H4C) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
8BVD
 
 | | FimH lectin domain in complex with mannose C-linked to quinoline | | Descriptor: | (2R,3S,4R,5S,6R)-2-(hydroxymethyl)-6-[(E)-3-quinolin-6-ylprop-2-enyl]oxane-3,4,5-triol, Type 1 fimbrin D-mannose specific adhesin | | Authors: | Bouckaert, J, Bridot, C. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Insightful Improvement in the Design of Potent Uropathogenic E. coli FimH Antagonists.
Pharmaceutics, 15, 2023
|
|
4VUB
 
 | | CCDB, A TOPOISOMERASE POISON FROM ESCHERICHIA COLI | | Descriptor: | CCDB, CHLORIDE ION | | Authors: | Loris, R, Dao-Thi, M.-H, Bahasi, E.M, Van Melderen, L, Poortmans, F, Liddington, R, Couturier, M, Wyns, L. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CcdB, a topoisomerase poison from E. coli.
J.Mol.Biol., 285, 1999
|
|
6ZIQ
 
 | | bovine ATP synthase stator domain, state 1 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZIT
 
 | | bovine ATP synthase Stator domain, state 2 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C2, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
