6NNE
 
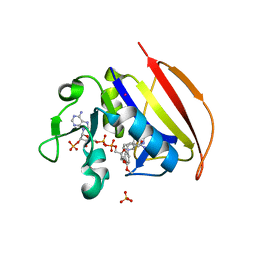 | | Crystal structure of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with diaverdine | | Descriptor: | 5-[(3,4-dimethoxyphenyl)methyl]pyrimidine-2,4-diamine, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5M24
 
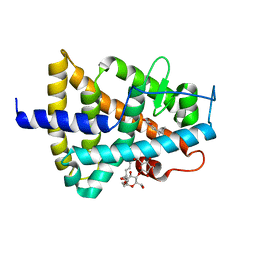 | | RARg mutant-S371E | | Descriptor: | (9cis)-retinoic acid, CHLORIDE ION, DODECYL-ALPHA-D-MALTOSIDE, ... | | Authors: | Rochel, N, Sirigu, S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Allosteric Regulation in the Ligand Binding Domain of Retinoic Acid Receptor gamma.
PLoS ONE, 12, 2017
|
|
5OTW
 
 | |
4PFN
 
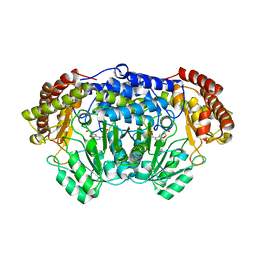 | | Crystal structure of Plasmodium vivax SHMT with L-serine Schiff base | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, SERINE, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2014-04-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5WIK
 
 | | JAK2 Pseudokinase in complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIM
 
 | | JAK2 Pseudokinase in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
2P1M
 
 | | TIR1-ASK1 complex structure | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, SKP1-like protein 1A, TRANSPORT INHIBITOR RESPONSE 1 protein | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-05 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
1OAN
 
 | |
4RXD
 
 | |
5AEN
 
 | | Structure of human Leukotriene A4 hydrolase in complex with inhibitor dimethyl(2- (4-phenoxyphenoxy)ethyl)amine | | Descriptor: | IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, N,N-dimethyl-2-(4-phenoxyphenoxy)ethanamine, ... | | Authors: | Moser, D, Wittmann, S.K, Kramer, J, Blocher, R, Achenbach, J, Pogoryelov, D, Proschak, E. | | Deposit date: | 2015-01-06 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Peng: A Neural Gas-Based Approach for Pharmacophore Elucidation. Method Design, Validation and Virtual Screening for Novel Ligands of Lta4H.
J.Chem.Inf.Model., 55, 2015
|
|
2GI4
 
 | |
6W1Q
 
 | | RT XFEL structure of Photosystem II 50 microseconds after the second illumination at 2.27 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5EFY
 
 | | Apo-form of SCO3201 | | Descriptor: | Putative tetR-family transcriptional regulator | | Authors: | Waack, P, Hinrichs, W. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of free and ligand-bound forms of the TetR/AcrR-like regulator SCO3201 from Streptomyces coelicolor suggest a novel allosteric mechanism.
Febs J., 2022
|
|
4RL3
 
 | | Crystal Structure of the Catalytic Domain of a family GH18 Chitinase from fern, Peteris ryukyuensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chitinase A, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A class III chitinase without disulfide bonds from the fern, Pteris ryukyuensis: crystal structure and ligand-binding studies.
Planta, 242, 2015
|
|
1VYD
 
 | | Crystal structure of cytochrome C2 mutant G95E | | Descriptor: | CYTOCHROME C2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dumortier, C, Fitch, J, Van Petegem, F, Vermeulen, W, Meyer, T.E, Van Beeumen, J.J, Cusanovich, M.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-06-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein Dynamics in the Region of the Sixth Ligand Methionine Revealed by Studies of Imidazole Binding to Rhodobacter Capsulatus Cytochrome C2 Hinge Mutants.
Biochemistry, 43, 2004
|
|
6W1T
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the second illumination at 2.01 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5VEC
 
 | |
2Q89
 
 | | Crystal structure of EhuB in complex with hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
6W1U
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4RXA
 
 | | Crystal structure of human farnesyl diphosphate synthase in complex with BPH-1358 | | Descriptor: | Farnesyl pyrophosphate synthase, N,N'-bis[3-(4,5-dihydro-1H-imidazol-2-yl)phenyl]biphenyl-4,4'-dicarboxamide, PHOSPHATE ION | | Authors: | Liu, Y.-L, Cao, R, Wang, Y, Oldfield, E. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Farnesyl diphosphate synthase inhibitors with unique ligand-binding geometries.
ACS Med Chem Lett, 6, 2015
|
|
5DDE
 
 | | Menin in complex with MI-859 | | Descriptor: | 1,2-ETHANEDIOL, 6-(2,2-difluoroethyl)-4-[4-(5,5-dimethyl-4,5-dihydro-1,3-thiazol-2-yl)piperazin-1-yl]thieno[2,3-d]pyrimidine, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Rational Design of Orthogonal Multipolar Interactions with Fluorine in Protein-Ligand Complexes.
J.Med.Chem., 58, 2015
|
|
4RYP
 
 | |
3VX7
 
 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | Descriptor: | E1, E2 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4BVN
 
 | | Ultra-thermostable beta1-adrenoceptor with cyanopindolol bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, ... | | Authors: | Miller, J, Nehme, R, Warne, T, Edwards, P.C, Leslie, A.G.W, Schertler, G, Tate, C.G. | | Deposit date: | 2013-06-26 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Resolution Structure of Cyanopindolol-Bound Beta1- Adrenoceptor Identifies an Intramembrane Na+ Ion that Stabilises the Ligand-Free Receptor.
Plos One, 9, 2014
|
|
9FQI
 
 | | E3 ligase Cbl-b in complex with a lactam scaffold inhibitor (compound 7) | | Descriptor: | 8-[3-[(4~{R})-4-methyl-2-oxidanylidene-piperidin-4-yl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
