1HHL
 
 | |
4RH5
 
 | |
4RH9
 
 | |
4RHG
 
 | |
5GSR
 
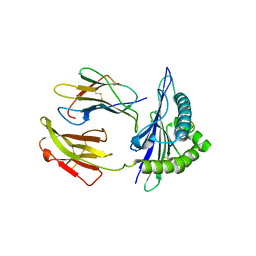 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide I5A | | Descriptor: | 9-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GR7
 
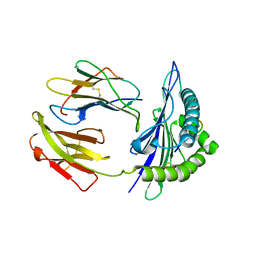 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-1 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5N9P
 
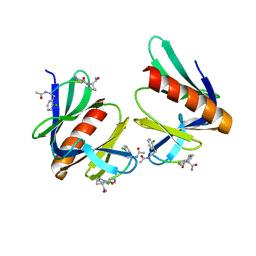 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-NH2 | | Descriptor: | Ac-[2-Cl-F]-PP-[ProM-1]-NH2, CHLORIDE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-26 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5GSB
 
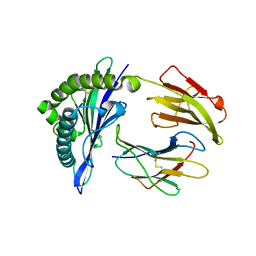 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-3 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-12 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5EJK
 
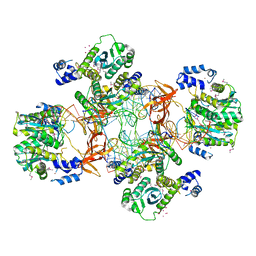 | | Crystal structure of the Rous sarcoma virus intasome | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*T)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*CP*TP*C)-3'), ... | | Authors: | Yin, Z, Shi, K, Banerjee, S, Aihara, H. | | Deposit date: | 2015-11-02 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of the Rous sarcoma virus intasome.
Nature, 530, 2016
|
|
5NMF
 
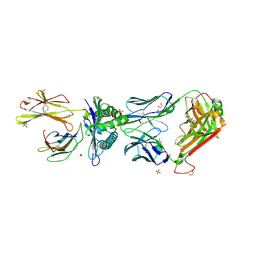 | | 868 TCR in complex with HLA A02 presenting SLYNTIATL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Dual Molecular Mechanisms Govern Escape at Immunodominant HLA A2-Restricted HIV Epitope.
Front Immunol, 8, 2017
|
|
8D8O
 
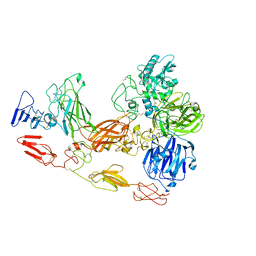 | | Cryo-EM structure of substrate unbound PAPP-A | | Descriptor: | Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
5D7W
 
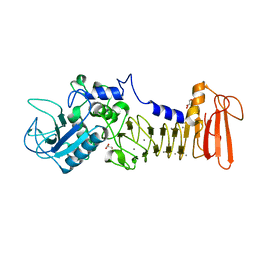 | | Crystal structure of serralysin | | Descriptor: | CALCIUM ION, GLYCEROL, Serralysin, ... | | Authors: | Wu, D, Ran, T, Xu, D.Q, Wang, W. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of a thermostable serralysin from Serratia sp. FS14 at 1.1 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5NI2
 
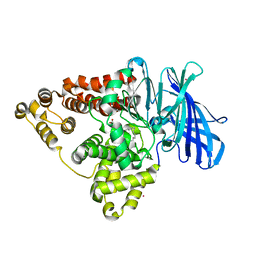 | |
5NC7
 
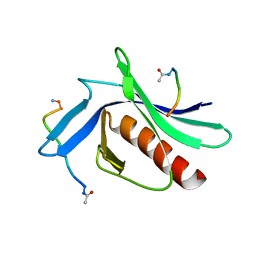 | | ENAH EVH1 in complex with Ac-WPPPPTEDEL-NH2 | | Descriptor: | ActA-derived 10-mer Ac-FPPPPTEDEL-NH2 with acetylated (Ac) and amidated (NH2) termini. Phe is substitued by Trp to increase affinity for crystallization, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-03 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5LKS
 
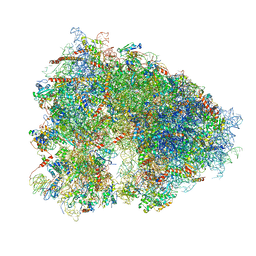 | | Structure-function insights reveal the human ribosome as a cancer target for antibiotics | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Myasnikov, A.G, Natchiar, S.K, Nebout, M, Hazemann, I, Imbert, V, Khatter, H, Peyron, J.-F, Klaholz, B.P. | | Deposit date: | 2016-07-23 | | Release date: | 2017-04-26 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-function insights reveal the human ribosome as a cancer target for antibiotics.
Nat Commun, 7, 2016
|
|
4SRN
 
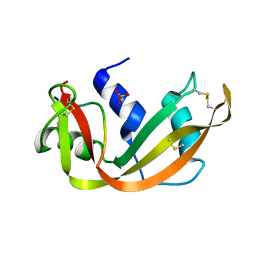 | | STRUCTURAL CHANGES THAT ACCOMPANY THE REDUCED CATALYTIC EFFICIENCY OF TWO SEMISYNTHETIC RIBONUCLEASE ANALOGS | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | deMel, V.S.J, Martin, P.D, Doscher, M.S, Edwards, B.F.P. | | Deposit date: | 1991-05-20 | | Release date: | 1994-12-20 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural changes that accompany the reduced catalytic efficiency of two semisynthetic ribonuclease analogs.
J.Biol.Chem., 267, 1992
|
|
8C9G
 
 | | Priestia megaterium mupirocin-sensitive isoleucyl-tRNA synthetase 1 complexed with mupirocin | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-22 | | Release date: | 2023-08-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8CZO
 
 | | Cryo-EM structure of BCL10 CARD - MALT1 DD filament | | Descriptor: | B-cell lymphoma/leukemia 10, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | David, L, Wu, H. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | BCL10 Mutations Define Distinct Dependencies Guiding Precision Therapy for DLBCL.
Cancer Discov, 12, 2022
|
|
2CEL
 
 | | ACTIVE-SITE MUTANT E212Q DETERMINED AT PH 6.0 WITH NO LIGAND BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
8F0V
 
 | | Lipocalin-like Milk protein-2 - E38A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein, ZINC ION | | Authors: | Subramanian, R, KanagaVijayan, D. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
8BXL
 
 | | Patulin Synthase from Penicillium expansum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tjallinks, G, Boverio, A, Rozeboom, H.J, Fraaije, M.W. | | Deposit date: | 2022-12-09 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure elucidation and characterization of patulin synthase, insights into the formation of a fungal mycotoxin.
Febs J., 290, 2023
|
|
8F0Y
 
 | | Lipocalin-like Milk protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein | | Authors: | Subramanian, R, KanagaVijayan, D, Shantakumar, R.P.S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
5G1X
 
 | | Crystal structure of Aurora-A kinase in complex with N-Myc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AURORA KINASE A, MAGNESIUM ION, ... | | Authors: | Richards, M.W, Burgess, S.G, Bayliss, R. | | Deposit date: | 2016-03-31 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis of N-Myc Binding by Aurora-A and its Destabilization by Kinase Inhibitors
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DAI
 
 | |
5NI4
 
 | |
