3MXY
 
 | |
2MMC
 
 | | Nucleotide-free human ran gtpase | | Descriptor: | GTP-binding nuclear protein Ran | | Authors: | Bacot-Davis, V.R, Palmenberg, A.C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of Ran GTPase Determines C-terminal Tail Conformational Dynamics.
To be Published
|
|
3MXD
 
 | | Crystal structure of HIV-1 protease inhibitor KC53 in complex with wild-type protease | | Descriptor: | (5S)-N-{(1S,2R)-3-[(1,3-benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-3-(2-hydroxyphenyl)-2 -oxo-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
3MVC
 
 | | High resolution crystal structure of the heme domain of GLB-6 from C. elegans | | Descriptor: | Globin protein 6, NITRATE ION, PRASEODYMIUM ION, ... | | Authors: | Yoon, J, Herzik Jr, M.A, Winter, M.B, Tran, R, Olea Jr, C, Marletta, M.A. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structure and properties of a bis-histidyl ligated globin from Caenorhabditis elegans.
Biochemistry, 49, 2010
|
|
3MVM
 
 | | P38 Alpha Map Kinase complexed with pyrrolotriazine inhibitor 7V | | Descriptor: | 4-{[5-(isoxazol-3-ylcarbamoyl)-2-methylphenyl]amino}-5-methyl-N-propylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-(5-(Cyclopropylcarbamoyl)-2-Methylphenylamino)-5-Methyl-Npropylpyrrolo[1,2-F][1,2,4] Triazine-6-Carboxamide (Bms-582949), a Clinical P38? Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
J.Med.Chem., 53, 2010
|
|
2M96
 
 | |
3MVL
 
 | | P38 Alpha Map Kinase complexed with pyrrolotriazine inhibitor 7K | | Descriptor: | 4-{[5-(cyclopropylcarbamoyl)-2-methylphenyl]amino}-5-methyl-N-propylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 4-(5-(Cyclopropylcarbamoyl)-2-Methylphenylamino)-5-Methyl-Npropylpyrrolo[1,2-F][1,2,4] Triazine-6-Carboxamide (Bms-582949), a Clinical P38 Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
J.Med.Chem., 53, 2010
|
|
3MW1
 
 | | p38 kinase Crystal structure in complex with small molecule inhibitor | | Descriptor: | 8-(2,6-dichlorophenyl)-4-(2,4-difluorophenyl)-2-piperidin-4-yl-1,7-naphthyridine 7-oxide, Mitogen-activated protein kinase 14 | | Authors: | Segarra, V, Caturla, F, Lumeras, W, Roca, R, Fisher, M, Lamers, M. | | Deposit date: | 2010-05-05 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 1,7-Naphthyridine 1-Oxides as Novel Potent and Selective Inhibitors of p38 Mitogen Activated Protein Kinase
J.Med.Chem., 54, 2011
|
|
3MVR
 
 | |
2MNG
 
 | | Apo Structure of human HCN4 CNBD solved by NMR | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Akimoto, M, Zhang, Z, Boulton, S, Selvaratnam, R, VanSchouwen, B, Gloyd, M, Accili, E.A, Lange, O.F, Melacini, G. | | Deposit date: | 2014-04-03 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP.
J.Biol.Chem., 289, 2014
|
|
3MWZ
 
 | |
3MWS
 
 | | Crystal Structure of Group N HIV-1 Protease | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease | | Authors: | Sayer, J.M, Agniswamy, J, Weber, I.T, Louis, J.M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: relevance to drug resistance.
Protein Sci., 19, 2010
|
|
3MX8
 
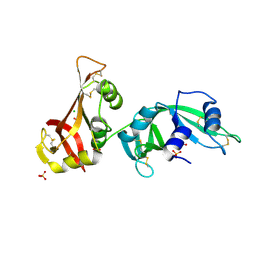 | | Crystal structure of ribonuclease A tandem enzymes and their interaction with the cytosolic ribonuclease inhibitor | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, LINKER, ... | | Authors: | Leich, F, Neumann, P, Lilie, H, Ulbrich-Hofmann, R, Arnold, U. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of RNase A tandem enzymes and their interaction with the cytosolic ribonuclease inhibitor
Febs J., 278, 2011
|
|
2LYZ
 
 | |
3MXH
 
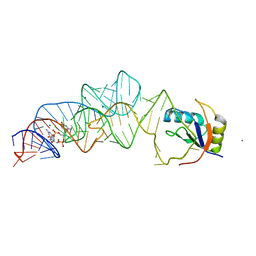 | | Native structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-07 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MYG
 
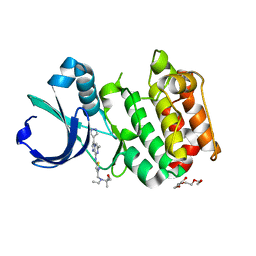 | | Aurora A Kinase complexed with SCH 1473759 | | Descriptor: | 2-{ethyl[(5-{[6-methyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}isothiazol-3-yl)methyl]amino}-2-methylpropan-1-ol, Serine/threonine-protein kinase 6, TETRAETHYLENE GLYCOL | | Authors: | Hruza, A, Prosis, W, Ramanathan, L. | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Injectable Inhibitor of Aurora Kinases Based on the Imidazo-[1,2-a]-Pyrazine Core.
ACS Med Chem Lett, 1, 2010
|
|
3MYN
 
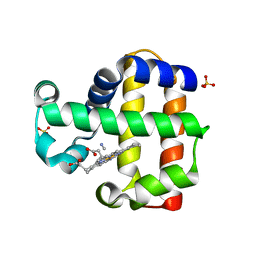 | | Mutation of Methionine-86 in Dehaloperoxidase-hemoglobin: Effects of the Asp-His-Fe Triad in a 3/3 Globin | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bowden, E.F, de Serrano, V.S, D'Antonio, E.L, Franzen, S. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Functional consequences of the creation of an Asp-His-Fe triad in a 3/3 globin.
Biochemistry, 50, 2011
|
|
3MTL
 
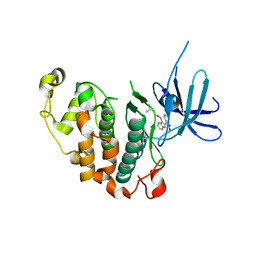 | | Crystal structure of the PCTAIRE1 kinase in complex with Indirubin E804 | | Descriptor: | (2Z,3E)-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE 3-{O-[(3R)-3,4-DIHYDROXYBUTYL]OXIME}, Cell division protein kinase 16 | | Authors: | Krojer, T, Sharpe, T.D, Roos, A, Savitsky, P, Amos, A, Ayinampudi, V, Berridge, G, Fedorov, O, Keates, T, Phillips, C, Burgess-Brown, N, Zhang, Y, Pike, A.C.W, Muniz, J, Vollmar, M, Thangaratnarajah, C, Rellos, P, Ugochukwu, E, Filippakopoulos, P, Yue, W, Das, S, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and inhibitor specificity of the PCTAIRE-family kinase CDK16.
Biochem.J., 474, 2017
|
|
2MY3
 
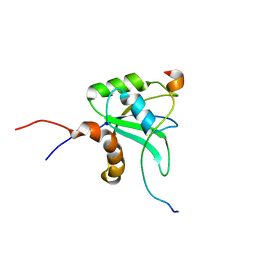 | |
3MWQ
 
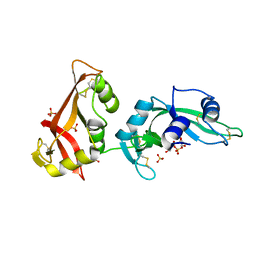 | |
3N3I
 
 | | Crystal Structure of G48V/C95F tethered HIV-1 Protease/Saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease | | Authors: | Prashar, V, Bihani, S.C, Das, A, Rao, D.R, Hosur, M.V. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of G48V/C95F tethered HIV-1 protease dimer/saquinavir complex
Biochem.Biophys.Res.Commun., 396, 2010
|
|
7C7J
 
 | |
3MU5
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2MP1
 
 | |
3MUT
 
 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
