7TII
 
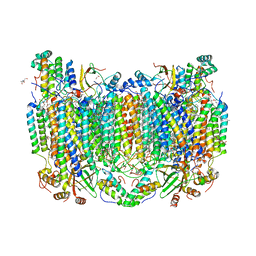 | | Annealed structure of oxidized bovine cytochrome c oxidase with reduced metal centers induced by synchrotron X-ray exposure | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Rousseau, D.L, Yeh, S.-R, Russi, S, Cohen, A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Temperature-dependent structural transition following X-ray-induced metal center reduction in oxidized cytochrome c oxidase.
J.Biol.Chem., 298, 2022
|
|
8OJH
 
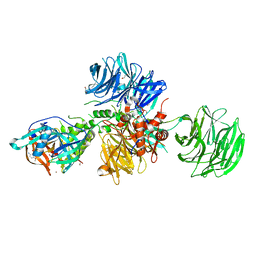 | | Crystal structure of human CRBN-DDB1 in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-2-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-7-methoxy-isoindole-1,3-dione, DNA damage-binding protein 1, ... | | Authors: | Cabry, M.P, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Chembiochem, 24, 2023
|
|
8P0M
 
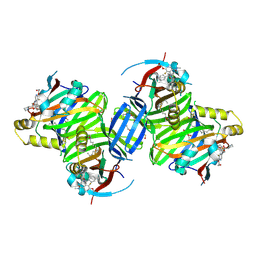 | | Crystal structure of TEAD3 in complex with IAG933 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-[(2~{S})-5-chloranyl-6-fluoranyl-2-phenyl-2-[(2~{S})-pyrrolidin-2-yl]-3~{H}-1-benzofuran-4-yl]-5-fluoranyl-6-(2-hydroxyethyloxy)-~{N}-methyl-pyridine-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Scheufler, C, Villard, F, Chau, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers.
Nat Cancer, 2024
|
|
7MJU
 
 | |
6WDZ
 
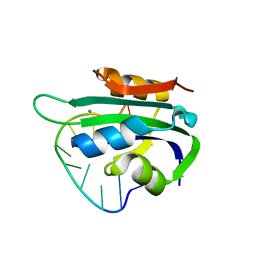 | | Porcine circovirus 2 Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent helicase Rep, DNA (5'-D(*TP*AP*GP*TP*AP*TP*TP*AP*CP*C)-3'), ... | | Authors: | Litzau, L.A, Tompkins, K, Shi, K, Nelson, A, Evans III, R.L, Gordon, W.R. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
6WKN
 
 | | PL-bound rat TRPV2 in nanodiscs | | Descriptor: | Transient receptor potential cation channel subfamily V member 2, piperlongumine | | Authors: | Pumroy, R.P, Moiseenkova-Bell, V.Y. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Allosteric Antagonist Modulation of TRPV2 by Piperlongumine Impairs Glioblastoma Progression.
Acs Cent.Sci., 7, 2021
|
|
8OUJ
 
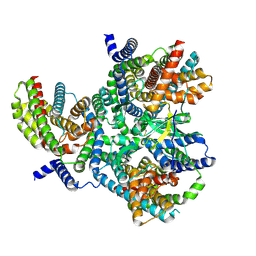 | | Heterotrimeric Complex of Human ASCT2 with Syncytin-1 | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Syncytin-1 | | Authors: | Khare, S, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 2024
|
|
7U0M
 
 | |
8OUI
 
 | | Complex of ASCT2 with Suppressyn | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Suppressyn | | Authors: | Khare, S, Kumar, A, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 2024
|
|
1H6H
 
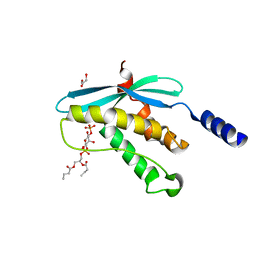 | | Structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, GLYCEROL, NEUTROPHIL CYTOSOL FACTOR 4 | | Authors: | Karathanassis, D, Bravo, J, Pacold, M, Perisic, O, Williams, R.L. | | Deposit date: | 2001-06-15 | | Release date: | 2001-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the Px Domain from P40Phox Bound to Phosphatidylinositol 3-Phosphate
Mol.Cell, 8, 2001
|
|
7TJQ
 
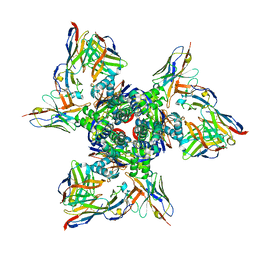 | | SAN27-14 bound to a antigenic site V on prefusion-stabilized hMPV F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, MPE8 Fab heavy chain, ... | | Authors: | Hsieh, C.-L, McLellan, J.S, Rush, S.A. | | Deposit date: | 2022-01-16 | | Release date: | 2022-09-14 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Characterization of prefusion-F-specific antibodies elicited by natural infection with human metapneumovirus.
Cell Rep, 40, 2022
|
|
7TL0
 
 | | Cryo-EM structure of hMPV preF bound by Fabs MPE8 and SAN32-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Rush, S.A, Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-09-14 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Characterization of prefusion-F-specific antibodies elicited by natural infection with human metapneumovirus.
Cell Rep, 40, 2022
|
|
7M5U
 
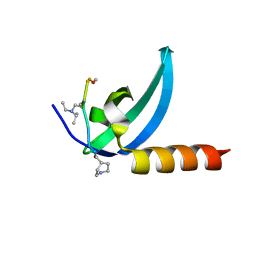 | | Crystal structure of human MPP8 chromodomain in complex with peptidomimetic ligand UNC5246 | | Descriptor: | M-phase phosphoprotein 8, UNC5246 | | Authors: | Budziszewski, G.R, McGinty, R.K, Waybright, J.M, Norris, J.L, James, L.I. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A Peptidomimetic Ligand Targeting the Chromodomain of MPP8 Reveals HRP2's Association with the HUSH Complex.
Acs Chem.Biol., 16, 2021
|
|
8OUH
 
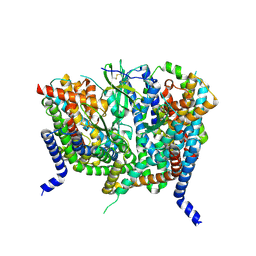 | | Complex of human ASCT2 with Syncytin-1 | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Syncytin-1 | | Authors: | Khare, S, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 2024
|
|
1KZ2
 
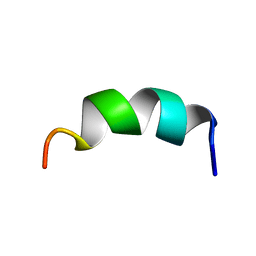 | | Solution structure of the third helix of Antennapedia homeodomain derivative [W6F,W14F] | | Descriptor: | Antennapedia protein | | Authors: | Czajlik, A, Mesko, E, Penke, B, Perczel, A. | | Deposit date: | 2002-02-06 | | Release date: | 2002-02-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of penetratin peptides. Part 1. The environment dependent conformational properties of penetratin and two of its derivatives.
J.Pept.Sci., 8, 2002
|
|
7UR1
 
 | |
7UM2
 
 | |
6XBI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW248 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XFN
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW243 | | Descriptor: | 3C-like proteinase, GLYCEROL, UAW243 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW247 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
1GQS
 
 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH NAP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(1S)-1-(DIMETHYLAMINO)ETHYL]PHENOL, ACETYLCHOLINESTERASE | | Authors: | Bar-on, P, Millard, C.B, Harel, M, Dvir, H, Enz, A, Sussman, J.L, Silman, I. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetic and Structural Studies on the Interaction of Cholinesterases with the Anti-Alzheimer Drug Rivastigmine
Biochemistry, 41, 2002
|
|
1H3Q
 
 | |
7LOP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV05-163 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab heavy chain, CR3022 Fab light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-10 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants.
Science, 373, 2021
|
|
1GU3
 
 | | CBM4 structure and function | | Descriptor: | ENDOGLUCANASE C, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
