2K3Y
 
 | |
2LSP
 
 | | solution structures of BRD4 second bromodomain with NF-kB-K310ac peptide | | Descriptor: | Bromodomain-containing protein 4, NF-kB-K310ac peptide | | Authors: | Zhang, G, Liu, R, Zhong, Y, Plotnikov, A.N, Zhang, W, Rusinova, E, Gerona-Nevarro, G, Moshkina, N, Joshua, J, Chuang, P.Y, Ohlmeyer, M, He, J, Zhou, M.-M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Down-regulation of NF-kappa B transcriptional activity in HIV-associated kidney disease by BRD4 inhibition.
J.Biol.Chem., 287, 2012
|
|
2L7F
 
 | |
2LFH
 
 | | Solution NMR Structure of the Helix-loop-Helix Domain of Human ID3 Protein, Northeast Structural Genomics Consortium Target HR3111A | | Descriptor: | DNA-binding protein inhibitor ID-3 | | Authors: | Eletsky, A, Wang, D, Kohan, E, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Helix-loop-Helix Domain of Human ID3 Protein, Northeast Structural Genomics Consortium Target HR3111A
To be Published
|
|
2LGD
 
 | |
2LGV
 
 | | Rbx1 | | Descriptor: | E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Spratt, D.E, Shaw, G.S. | | Deposit date: | 2011-08-02 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective recruitment of an e2~ubiquitin complex by an e3 ubiquitin ligase.
J.Biol.Chem., 287, 2012
|
|
2LF8
 
 | |
2KO3
 
 | | Nedd8 solution structure | | Descriptor: | NEDD8 | | Authors: | Choi, Y.S, Jeon, Y.H, Cheong, C. | | Deposit date: | 2009-09-09 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 60th residues of ubiquitin and Nedd8 are located out of E2-binding surfaces, but are important for K48 ubiquitin-linkage.
Febs Lett., 583, 2009
|
|
2KR0
 
 | |
2KBM
 
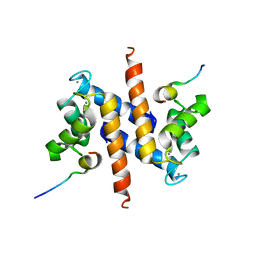 | | Ca-S100A1 interacting with TRTK12 | | Descriptor: | CALCIUM ION, F-actin-capping protein subunit alpha-2, Protein S100-A1 | | Authors: | Wright, N.T, Varney, K.M, Cannon, B.R, Morgan, M, Weber, D.J. | | Deposit date: | 2008-12-02 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ca-S100A1-TRTK12
To be Published
|
|
2ME6
 
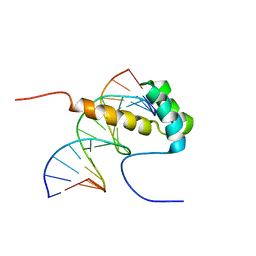 | | NMR Structure of the homeodomain transcription factor Gbx1 from Homo sapiens in complex with the DNA sequence CGACTAATTAGTCG | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*TP*AP*AP*TP*TP*AP*GP*TP*CP*G)-3'), Homeobox protein GBX-1 | | Authors: | Proudfoot, A, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the homeodomain transcription factor Gbx1 from Homo sapiens in complex with the DNA sequence CGACTAATTAGTCG
To be Published
|
|
2MGQ
 
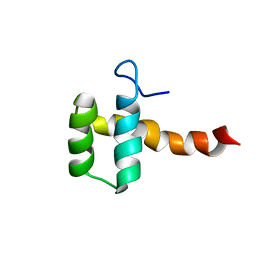 | | Structure of CEH37 Homeodomain | | Descriptor: | Homeobox protein ceh-37 | | Authors: | Moon, S, Lee, W. | | Deposit date: | 2013-11-04 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of CEH37 Homeodomain
To be Published
|
|
2M0C
 
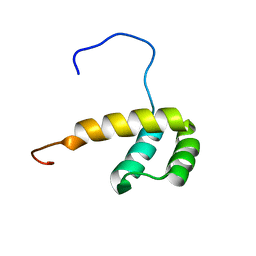 | | Solution NMR Structure of Homeobox Domain of Human ALX4, Northeast Structural Genomics Consortium (NESG) Target HR4490C | | Descriptor: | Homeobox protein aristaless-like 4 | | Authors: | Xu, X, Eletsky, A, Pulavarti, S, Lee, D, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-24 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Homeobox Domain of Human ALX4, Northeast Structural Genomics Consortium (NESG) Target HR4490C
To be Published
|
|
2L9R
 
 | | Solution NMR Structure of Homeobox domain of Homeobox protein Nkx-3.1 from homo sapiens, Northeast Structural Genomics Consortium Target HR6470A | | Descriptor: | Homeobox protein Nkx-3.1 | | Authors: | Liu, G, Xiao, R, Lee, H.-W, Hamilton, K, Ciccosanti, C, Wang, H.B, Acton, T.B, Everett, J.K, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Homeobox domain of Homeobox protein Nkx-3.1 from homo sapiens, Northeast Structural Genomics Consortium Target HR6470A
To be Published
|
|
2K3X
 
 | | Solution structure of EAF3 chromo barrel domain | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Mer, G, Xu, C. | | Deposit date: | 2008-05-19 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Recognition of Methylated Histone H3K36 by the Eaf3 Subunit of Histone Deacetylase Complex Rpd3S.
Structure, 16, 2008
|
|
2KK0
 
 | | Solution structure of dead ringer-like protein 1 (at-rich interactive domain-containing protein 3a) from homo sapiens, northeast structural genomics consortium (NESG) target hr4394c | | Descriptor: | AT-rich interactive domain-containing protein 3A | | Authors: | Liu, G, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-14 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ARID domain of human AT-rich interactive domain-containing protein 3A: a human cancer protein interaction network target.
Proteins, 78, 2010
|
|
2M34
 
 | |
2MD5
 
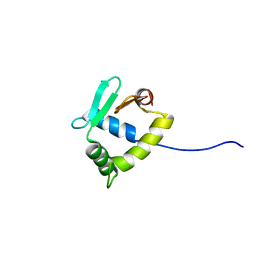 | | Structure of uninhibited ETV6 ETS domain | | Descriptor: | Transcription factor ETV6 | | Authors: | De, S, Mcintosh, L.P, Chan, A.C, Coyne, H.J, Okon, M, Graves, B.J, Murphy, M.E. | | Deposit date: | 2013-08-29 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
5I42
 
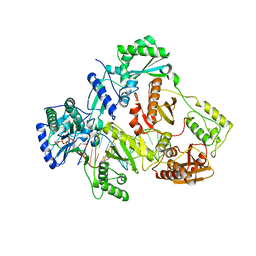 | | Structure of HIV-1 Reverse Transcriptase in complex with a DNA aptamer, AZTTP, and CA(2+) ion | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (38-MER), ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet.
Acs Chem.Biol., 11, 2016
|
|
5HP1
 
 | |
5I3U
 
 | |
3MRI
 
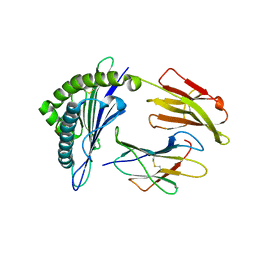 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide G4M-V5W variant | | Descriptor: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Reiser, J.-B, Le Gorrec, M, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide G4M-V5W variant
To be Published
|
|
3MRL
 
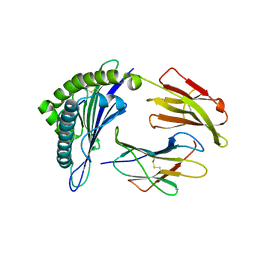 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide C6V variant | | Descriptor: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Reiser, J.-B, Chouquet, A, Le Gorrec, M, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
3MRJ
 
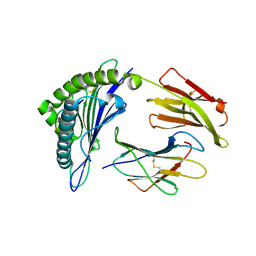 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide V5M variant | | Descriptor: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Reiser, J.-B, Le Gorrec, M, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide V5M variant
To be Published
|
|
6K3L
 
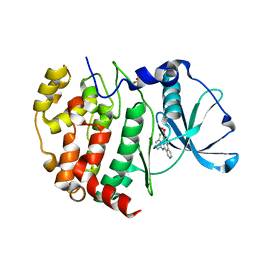 | | Crystal structure of CX-4945 bound Cka1 from C. neoformans | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CMGC/CK2 protein kinase, SULFATE ION | | Authors: | Cho, H.S, Yoo, Y. | | Deposit date: | 2019-05-20 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural analysis of fungal pathogenicity-related casein kinase alpha subunit, Cka1, in the human fungal pathogen Cryptococcus neoformans.
Sci Rep, 9, 2019
|
|
