7YPV
 
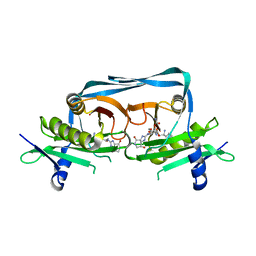 | | Crystal structure of OrE-ST-F | | Descriptor: | Acetyltransferase, Streptothricin F | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-08-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
3B1M
 
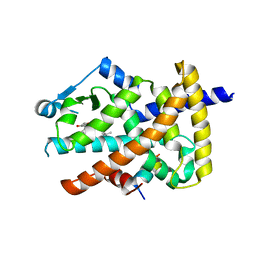 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator Cerco-A | | Descriptor: | (9aS)-8-acetyl-N-[(2-ethylnaphthalen-1-yl)methyl]-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Matsui, Y, Hiroyuki, H. | | Deposit date: | 2011-07-05 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pharmacology and in Vitro Profiling of a Novel Peroxisome Proliferator-Activated Receptor gamma Ligand, Cerco-A
Biol.Pharm.Bull., 34, 2011
|
|
8OKX
 
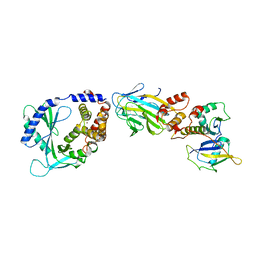 | | Structure of cGAS in complex with SPSB3-ELOBC | | Descriptor: | Cyclic GMP-AMP synthase, Elongin-B, Elongin-C, ... | | Authors: | Xu, P.B, Ablasser, A. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | The CRL5-SPSB3 ubiquitin ligase targets nuclear cGAS for degradation.
Nature, 627, 2024
|
|
1CUO
 
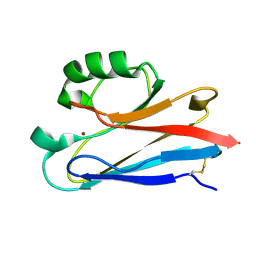 | | CRYSTAL STRUCTURE ANALYSIS OF ISOMER-2 AZURIN FROM METHYLOMONAS J | | Descriptor: | COPPER (II) ION, PROTEIN (AZURIN ISO-2) | | Authors: | Inoue, T, Nishio, N, Kai, Y, Suzuki, S, Kataoka, K. | | Deposit date: | 1999-08-21 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J.
J.Mol.Biol., 333, 2003
|
|
8OL1
 
 | |
5LSV
 
 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5M2V
 
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with (2S,4R)-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid at 3.18 A resolution | | Descriptor: | (2~{S},4~{R})-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Frydenvang, K, Kastrup, J.S, Kristensen, C.M. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Design and Synthesis of a Series of l-trans-4-Substituted Prolines as Selective Antagonists for the Ionotropic Glutamate Receptors Including Functional and X-ray Crystallographic Studies of New Subtype Selective Kainic Acid Receptor Subtype 1 (GluK1) Antagonist (2S,4R)-4-(2-Carboxyphenoxy)pyrrolidine-2-carboxylic Acid.
J. Med. Chem., 60, 2017
|
|
5M5F
 
 | | Thermolysin in complex with inhibitor and krypton | | Descriptor: | (2~{S})-4-methyl-2-[2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]ethanoylamino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-10-21 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
5IRZ
 
 | | Structure of TRPV1 determined in lipid nanodisc | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4S,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(pentanoyloxy)propan-2-yl decanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, (4R,7S)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(pentanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphatetradecan-1-aminium, ... | | Authors: | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2016-03-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
1ZHB
 
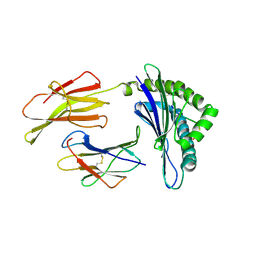 | | Crystal Structure Of The Murine Class I Major Histocompatibility Complex Of H-2Db, B2-Microglobulin, and a 9-Residue Peptide Derived from rat dopamine beta-monooxigenase | | Descriptor: | 9-mer peptide from Dopamine beta-monooxygenase, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Sandalova, T, Michaelsson, J, Harris, R.A, Odeberg, J, Schneider, G, Karre, K, Achour, A. | | Deposit date: | 2005-04-25 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural basis for CD8+ T cell-dependent recognition of non-homologous peptide ligands: implications for molecular mimicry in autoreactivity
J.Biol.Chem., 280, 2005
|
|
5MA7
 
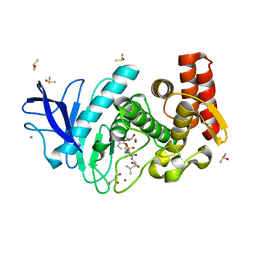 | | Structure of thermolysin in complex with inhibitor (JC306). | | Descriptor: | (2~{S})-2-[[(2~{S})-3-azanyl-2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]propanoyl]amino]-4-methyl-pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-11-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
6ZB2
 
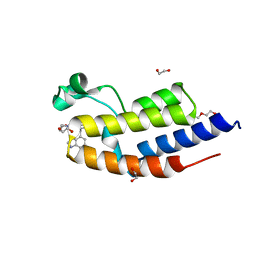 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH GSK549 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chung, C. | | Deposit date: | 2020-06-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor.
J.Med.Chem., 63, 2020
|
|
7QRW
 
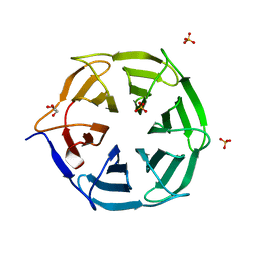 | |
5M69
 
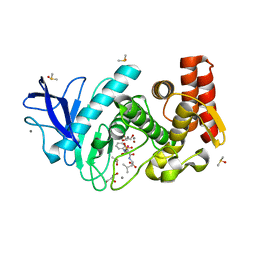 | | Thermolysin in complex with inhibitor and xenon | | Descriptor: | (2~{S})-4-methyl-2-[2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]ethanoylamino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-10-24 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
5MNG
 
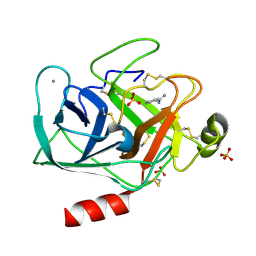 | | Cationic trypsin in complex with benzamidine (deuterated sample at 100 K) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
5MNZ
 
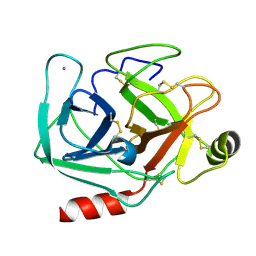 | | Neutron structure of cationic trypsin in its apo form | | Descriptor: | CALCIUM ION, Cationic trypsin | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (1.45 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
5MOS
 
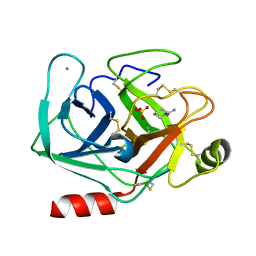 | | Joint X-ray/neutron structure of cationic trypsin in complex with N-amidinopiperidine | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-13 | | Method: | NEUTRON DIFFRACTION (0.96 Å), X-RAY DIFFRACTION | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
5LVD
 
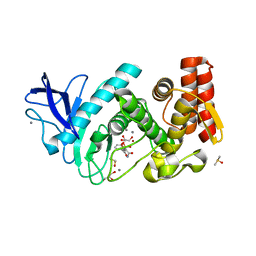 | | Thermolysin in complex with inhibitor (JC67) | | Descriptor: | (2~{S})-4-methyl-2-[[(2~{S})-3-oxidanyl-2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]propanoyl]amino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-14 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
5MNH
 
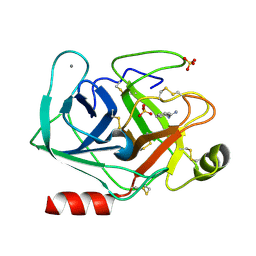 | | Cationic trypsin in complex with benzamidine (deuterated sample at 295 K) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
5MNO
 
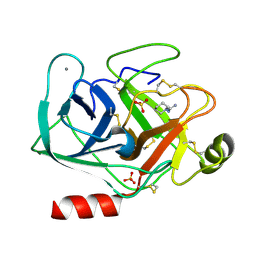 | | Cationic trypsin in complex with N-amidinopiperidine (deuterated sample at 295 K) | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
5MOQ
 
 | | Joint X-ray/neutron structure of cationic trypsin in complex with benzamidine | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-20 | | Method: | NEUTRON DIFFRACTION (0.93 Å), X-RAY DIFFRACTION | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
6E3S
 
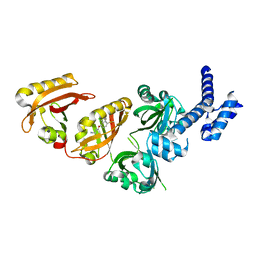 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist PT2385 | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6E3T
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist T1001 | | Descriptor: | (6S)-6-(4-bromophenyl)-2,3,5,6-tetrahydroimidazo[2,1-b][1,3]thiazole, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6E3U
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Agonist M1001 | | Descriptor: | 3-{[2-(pyrrolidin-1-yl)phenyl]amino}-1H-1lambda~6~,2-benzothiazole-1,1-dione, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
7W8Z
 
 | |
