7L2W
 
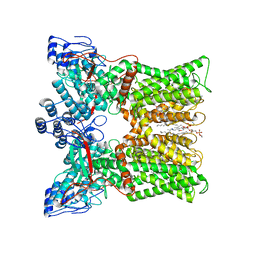 | | cryo-EM structure of RTX-bound minimal TRPV1 with NMDG at state a | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2S
 
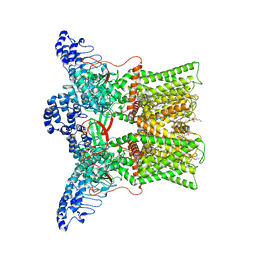 | | cryo-EM structure of DkTx-bound minimal TRPV1 at the pre-bound state | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2T
 
 | | cryo-EM structure of DkTx-bound minimal TRPV1 in partial open state | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, (9R,12R)-15-amino-12-hydroxy-6,12-dioxo-7,11,13-trioxa-12lambda~5~-phosphapentadecan-9-yl undecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2P
 
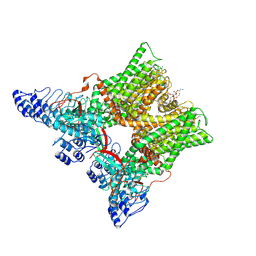 | | cryo-EM structure of unliganded minimal TRPV1 | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2X
 
 | | cryo-EM structure of RTX-bound minimal TRPV1 with NMDG at state c | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, 1-deoxy-1-(methylamino)-D-allitol, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L26
 
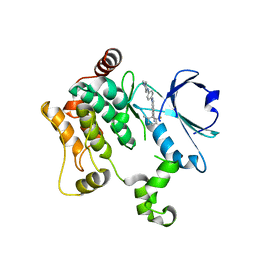 | | HPK1 IN COMPLEX WITH COMPOUND 38 | | Descriptor: | 6-(2-fluoro-6-methylphenyl)-1-[4-(4-methylpiperazin-1-yl)phenyl]-1H-indazole-5-carbonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Potent Reverse Indazole Inhibitors for HPK1.
Acs Med.Chem.Lett., 12, 2021
|
|
7L25
 
 | | HPK1 IN COMPLEX WITH COMPOUND 18 | | Descriptor: | 1,2-ETHANEDIOL, 6-(2-fluoro-6-methoxyphenyl)-1-[6-(4-methylpiperazin-1-yl)pyridin-2-yl]-1H-pyrazolo[4,3-c]pyridine, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of Potent Reverse Indazole Inhibitors for HPK1.
Acs Med.Chem.Lett., 12, 2021
|
|
7L24
 
 | | HPK1 IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 6-(2-fluoro-6-methoxyphenyl)-1-[4-(4-methylpiperazin-1-yl)phenyl]-1H-pyrazolo[4,3-c]pyridine, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Identification of Potent Reverse Indazole Inhibitors for HPK1.
Acs Med.Chem.Lett., 12, 2021
|
|
7BB1
 
 | | Lysozyme crystallized in the presence of the hydrated deep eutectic solvent Choline chloride-Glutamic acid 2:1 | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Belviso, B.D, Caliandro, R, Carrozzini, B. | | Deposit date: | 2020-12-16 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Introducing Protein Crystallization in Hydrated Deep Eutectic Solvents
Acs Sustain Chem Eng, 9, 2021
|
|
7BAZ
 
 | | Lysozyme crystallized in the presence of the hydrated deep eutectic solvent Choline chloride-Glycerol 1:2 | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Belviso, B.D, Caliandro, R, Carrozzini, B. | | Deposit date: | 2020-12-16 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Introducing Protein Crystallization in Hydrated Deep Eutectic Solvents
Acs Sustain Chem Eng, 9, 2021
|
|
7BAQ
 
 | |
7BAR
 
 | |
7BB0
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with NAT22-366511 and PKI (5-24) | | Descriptor: | (R)-3,3-dimethyl-5-oxo-5-(6-(3-(pyridin-4-yl)-1,2,4-oxadiazol-5-yl)-1,4,6,7-tetrahydro-5H-imidazo[4,5-c]pyridin-5-yl)pentanoic acid, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Oebbeke, M, Mueller, J, Glinca, S, Heine, A, Klebe, G. | | Deposit date: | 2020-12-16 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with NAT22-366511 and PKI (5-24)
To Be Published
|
|
7L1S
 
 | | PS3 F1-ATPase Pi-bound Dwell | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7L1Q
 
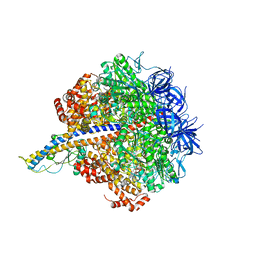 | | PS3 F1-ATPase Binding/TS Dwell | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7L1R
 
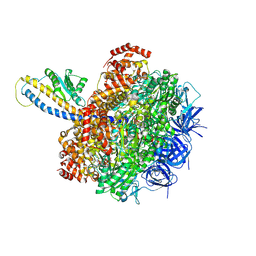 | | PS3 F1-ATPase Hydrolysis Dwell | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7L1X
 
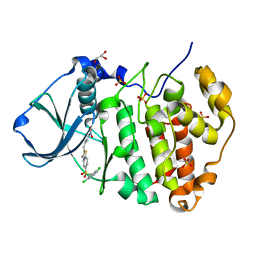 | | Structure of human CK2 alpha kinase (catalytic subunit) with the inhibitor 108600. | | Descriptor: | (2~{Z})-6-[[2,6-bis(chloranyl)phenyl]methylsulfonyl]-2-[[4-oxidanyl-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]methylidene]-4~{H}-1,4-benzothiazin-3-one, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Rechkoblit, O, Aggarwal, A.K. | | Deposit date: | 2020-12-15 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Simultaneous CK2/TNIK/DYRK1 inhibition by 108600 suppresses triple negative breast cancer stem cells and chemotherapy-resistant disease.
Nat Commun, 12, 2021
|
|
7L20
 
 | |
7L1U
 
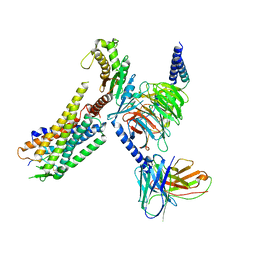 | | Orexin Receptor 2 (OX2R) in Complex with G Protein and Natural Peptide-Agonist Orexin B (OxB) | | Descriptor: | Engineered Guanine nucleotide-binding protein subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Hong, C, Byrne, N.J, Zamlynny, B, Tummala, S, Xiao, L, Shipman, J.M, Partridge, A.T, Minnick, C, Breslin, M.J, Rudd, M.T, Stachel, S.J, Rada, V.L, Kern, J.C, Armacost, K.A, Hollingsworth, S.A, O'Brien, J.A, Hall, D.L, McDonald, T.P, Strickland, C, Brooun, A, Soisson, S.M, Hollenstein, K. | | Deposit date: | 2020-12-15 | | Release date: | 2021-02-10 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation.
Nat Commun, 12, 2021
|
|
7L1V
 
 | | Orexin Receptor 2 (OX2R) in Complex with G Protein and Small-Molecule Agonist Compound 1 | | Descriptor: | 4'-methoxy-N,N-dimethyl-3'-{[3-(2-{[2-(2H-1,2,3-triazol-2-yl)benzene-1-carbonyl]amino}ethyl)phenyl]sulfamoyl}[1,1'-biphenyl]-3-carboxamide, Engineered Guanine nucleotide-binding protein subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hong, C, Byrne, N.J, Zamlynny, B, Tummala, S, Xiao, L, Shipman, J.M, Partridge, A.T, Minnick, C, Breslin, M.J, Rudd, M.T, Stachel, S.J, Rada, V.L, Kern, J.C, Armacost, K.A, Hollingsworth, S.A, O'Brien, J.A, Hall, D.L, McDonald, T.P, Strickland, C, Brooun, A, Soisson, S.M, Hollenstein, K. | | Deposit date: | 2020-12-15 | | Release date: | 2021-02-10 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation.
Nat Commun, 12, 2021
|
|
7B9J
 
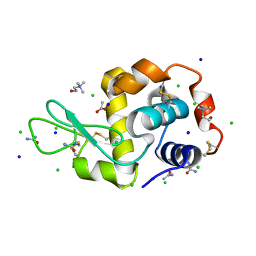 | | Lysozyme crystallized in the presence of the hydrated deep eutectic solvent Choline chloride-Urea 1:2 | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Belviso, B.D, Caliandro, R, Carrozzini, B. | | Deposit date: | 2020-12-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Introducing Protein Crystallization in Hydrated Deep Eutectic Solvents
Acs Sustain Chem Eng, 9, 2021
|
|
7B9L
 
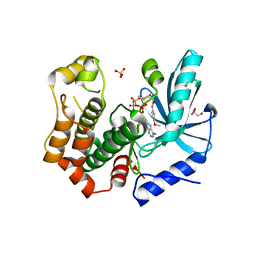 | | MEK1 in complex with compound 23 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7B94
 
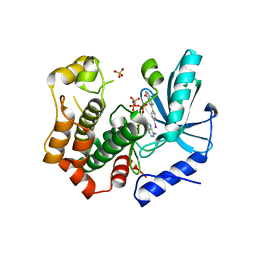 | | MEK1 in complex with compound 6 | | Descriptor: | 2-(4-iodophenyl)-8~{H}-imidazo[1,2-c]pyrimidin-5-one, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7B9V
 
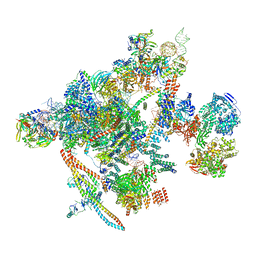 | | Yeast C complex spliceosome at 2.8 Angstrom resolution with Prp18/Slu7 bound | | Descriptor: | 5' exon of UBC4 mRNA, BJ4_G0027490.mRNA.1.CDS.1, BJ4_G0054360.mRNA.1.CDS.1, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Nagai, K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for conformational equilibrium of the catalytic spliceosome.
Mol.Cell, 81, 2021
|
|
7B7Z
 
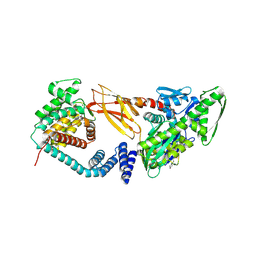 | | DeAMPylation complex of monomeric FICD and AMPylated BiP (state 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Endoplasmic reticulum chaperone BiP, ... | | Authors: | Perera, L.A, Ron, D. | | Deposit date: | 2020-12-12 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of a deAMPylation complex rationalise the switch between antagonistic catalytic activities of FICD.
Nat Commun, 12, 2021
|
|
