4ZZZ
 
 | | Structure of human PARP1 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-(3-methoxypropyl)-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, GLYCEROL, POLY [ADP-RIBOSE] POLYMERASE 1, ... | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
7Z8S
 
 | | Mot1:TBP:DNA - post hydrolysis state | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z7N
 
 | | Mot1E1434Q:TBP:DNA - substrate recognition state | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZB5
 
 | | Mot1(1-1836):TBP:DNA - post-hydrolysis complex dimer | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5MHC
 
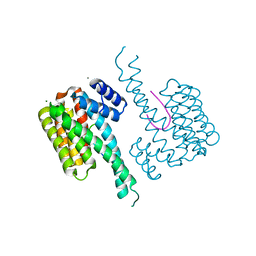 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, LYS-LEU-MET-PHE-LYS-TPO-GLU-GLY-PRO-ASP-SER-ASP, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2016-11-24 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
8CDG
 
 | | Crystal structure of human IL-17A cytokine in complex with macrocycle | | Descriptor: | (9~{S},12~{R},19~{S})-9-[[4-[[(2~{S})-2-cyclohexyl-2-(2-phenylethanoylamino)ethanoyl]amino]phenyl]methyl]-12-methyl-7,10,13,21-tetrakis(oxidanylidene)-8,11,14,20-tetrazaspiro[4.17]docosane-19-carboxylic acid, Interleukin-17A | | Authors: | Schulze, M.-S. | | Deposit date: | 2023-01-30 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Modulation of IL-17 backbone dynamics reduces receptor affinity and reveals a new inhibitory mechanism.
Chem Sci, 14, 2023
|
|
3QYW
 
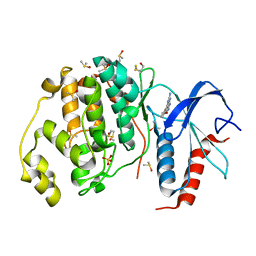 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 6-(3-bromophenyl)-7H-purin-2-amine, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3VKW
 
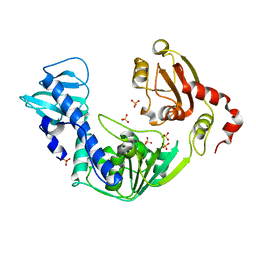 | | Crystal Structure of the Superfamily 1 Helicase from Tomato Mosaic Virus | | Descriptor: | Replicase large subunit, SULFATE ION | | Authors: | Nishikiori, M, Sugiyama, S, Xiang, H, Niiyama, M, Ishibashi, K, Inoue, T, Ishikawa, M, Matsumura, H, Katoh, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superfamily 1 helicase from tomato mosaic virus
J.Virol., 86, 2012
|
|
5MOC
 
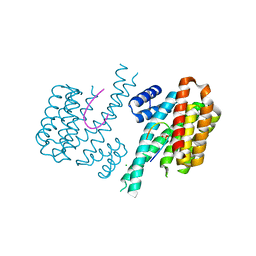 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2016-12-14 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
5NSD
 
 | | Co-crystal structure of NAMPT dimer with KPT-9274 | | Descriptor: | (~{E})-3-(6-azanylpyridin-3-yl)-~{N}-[[5-[4-[4,4-bis(fluoranyl)piperidin-1-yl]carbonylphenyl]-7-(4-fluorophenyl)-1-benzofuran-2-yl]methyl]prop-2-enamide, GLYCEROL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Neggers, J.E, Kwanten, B, Dierckx, T, Noguchi, H, Voet, A, Vercruysse, T, Baloglu, E, Senapedis, W, Jacquemyn, M, Daelemans, D. | | Deposit date: | 2017-04-26 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes.
Nat Commun, 9, 2018
|
|
6VCD
 
 | | Cryo-EM structure of IRP2-FBXL5-SKP1 complex | | Descriptor: | F-box/LRR-repeat protein 5, FE2/S2 (INORGANIC) CLUSTER, Iron-responsive element binding protein 2, ... | | Authors: | Wang, H, Shi, H, Zheng, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster.
Mol.Cell, 78, 2020
|
|
1U9L
 
 | | Structural basis for a NusA- protein N interaction | | Descriptor: | GOLD ION, Lambda N, Transcription elongation protein nusA | | Authors: | Bonin, I, Muehlberger, R, Bourenkov, G.P, Huber, R, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-08-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the interaction of Escherichia coli NusA with protein N of phage lambda
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
7ZKE
 
 | | Mot1:TBP:DNA - pre-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (36-MER), ... | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1JBP
 
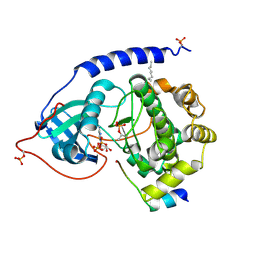 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Substrate Peptide, ADP and Detergent | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, MUSCLE/BRAIN FORM, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-06-06 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
1JLU
 
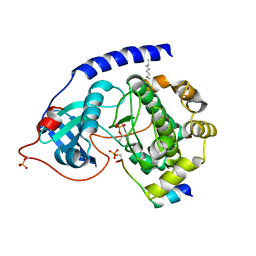 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Phosphorylated Substrate Peptide and Detergent | | Descriptor: | AMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.-H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
3QYZ
 
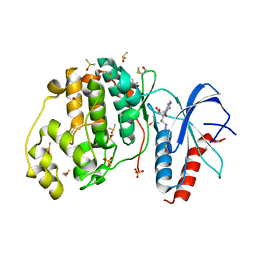 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 5'-azido-8-bromo-5'-deoxyadenosine, BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4JZN
 
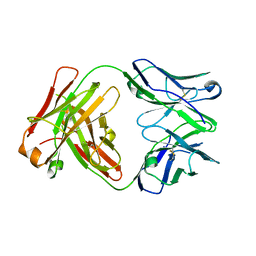 | |
4JZO
 
 | |
3VKX
 
 | | Structure of PCNA | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, CHLORIDE ION, Proliferating cell nuclear antigen, ... | | Authors: | Hashimoto, H, Hishiki, A, Shimizu, T, Sato, M, Punchihewa, C, Connelly, M, Actis, M, Waddell, B, Pagala, V, Fujii, N. | | Deposit date: | 2011-11-26 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication
J.Biol.Chem., 287, 2012
|
|
3F2D
 
 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP, Mn and Zn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1NHA
 
 | | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP-Binding Sites of RAP74 and CTD of RAP74, the subunit of Human TFIIF | | Descriptor: | Transcription initiation factor IIF, alpha subunit | | Authors: | Nguyen, B.D, Chen, H.T, Kobor, M.S, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP1-Binding Sites of RAP74 and Human TFIIB.
Biochemistry, 42, 2003
|
|
3QWW
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with the methyltransferase inhibitor sinefungin | | Descriptor: | SET and MYND domain-containing protein 2, SINEFUNGIN, ZINC ION | | Authors: | Jiang, Y, Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2011-02-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of histone and p53 methyltransferase SmyD2 reveal a conformational flexibility of the autoinhibitory C-terminal domain.
Plos One, 6, 2011
|
|
1RYR
 
 | | REPLICATION OF A CIS-SYN THYMINE DIMER AT ATOMIC RESOLUTION | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3', 5'-D(*TP*TP*TP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ling, H, Boudsocq, F, Plosky, B, Woodgate, R, Yang, W. | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Replication of a Cis-Syn Thymine Dimer at Atomic Resolution
Nature, 424, 2003
|
|
8FWL
 
 | |
3QWV
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with the cofactor product AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 2, ZINC ION | | Authors: | Jiang, Y, Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2011-02-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of histone and p53 methyltransferase SmyD2 reveal a conformational flexibility of the autoinhibitory C-terminal domain.
Plos One, 6, 2011
|
|
