5EO3
 
 | |
5EOC
 
 | | Crystal structure of Fab C2 in complex with a Cyclic variant of Hepatitis C Virus E2 epitope I | | Descriptor: | ALA-CYS-GLN-LEU-ILE-ASN-THR-ASN-GLY-SER-TRP-HIS-ILE-CYS, Fab fragment (Heavy chain), Fab fragment (Light chain) | | Authors: | Berisio, R, Ruggiero, A, Sandomenico, A, Patel, A.H, Ruvo, M, Vitagliano, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generation and Characterization of Monoclonal Antibodies against a Cyclic Variant of Hepatitis C Virus E2 Epitope 412-422.
J.Virol., 90, 2016
|
|
6XIN
 
 | | The crystal structure of tryptophan synthase from Salmonella enterica serovar typhimurium in complex with (2S)-3-Amino-3-imino-2-phenyldiazenylpropanamide at the enzyme alpha-site. | | Descriptor: | (2R,3Z)-3-amino-3-imino-2-[(E)-phenyldiazenyl]propanamide, 1,2-ETHANEDIOL, CESIUM ION, ... | | Authors: | Hilario, E, Chang, C, Mueller, L.J, Dunn, M.F, Fan, L. | | Deposit date: | 2020-06-20 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of tryptophan synthase from Salmonella enterica serovar typhimurium in complex with (2S)-3-Amino-3-imino-2-phenyldiazenylpropanamide at the enzyme alpha-site.
To Be Published
|
|
2W17
 
 | | CDK2 in complex with the imidazole pyrimidine amide, compound (S)-8b | | Descriptor: | ACETATE ION, CELL DIVISION PROTEIN KINASE 2, N-(4-{[(3S)-3-(dimethylamino)pyrrolidin-1-yl]carbonyl}phenyl)-5-fluoro-4-[2-methyl-1-(1-methylethyl)-1H-imidazol-5-yl]pyrimidin-2-amine | | Authors: | Jones, C.D, Andrews, D.M, Barker, A.J, Blades, K, Daunt, P, East, S, Geh, C, Graham, M.A, Johnson, K.M, Loddick, S.A, McFarland, H.M, McGregor, A, Moss, L, Rudge, D.A, Simpson, P.B, Swain, M.L, Tam, K.Y, Tucker, J.A, Walker, M, Brassington, C, Haye, H, McCall, E. | | Deposit date: | 2008-10-15 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Discovery of Azd5597, a Potent Imidazole Pyrimidine Amide Cdk Inhibitor Suitable for Intravenous Dosing.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VOJ
 
 | | Ternary complex of M. tuberculosis Rv2780 with NAD and pyruvate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, ALANINE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-02-18 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Mycobacterium Tuberculosis Secretory Antigen Alanine Dehydrogenase (Rv2780) in Apo and Ternary Complex Forms Captures "Open" and "Closed" Enzyme Conformations.
Proteins: Struct., Funct., Bioinf., 72, 2008
|
|
5F24
 
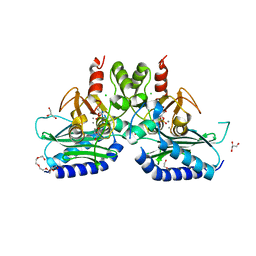 | | Crystal structure of dual specific IMPase/NADP phosphatase bound with D-inositol-1-phosphate | | Descriptor: | CALCIUM ION, CHLORIDE ION, D-MYO-INOSITOL-1-PHOSPHATE, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EYH
 
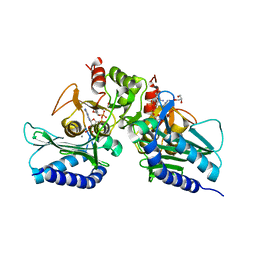 | | Crystal Structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ at pH 7.0 | | Descriptor: | CALCIUM ION, GLYCEROL, Inositol monophosphatase, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EYG
 
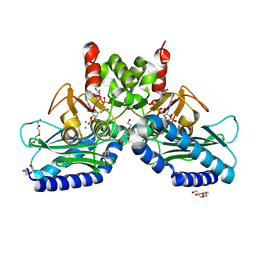 | | Crystal structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EW1
 
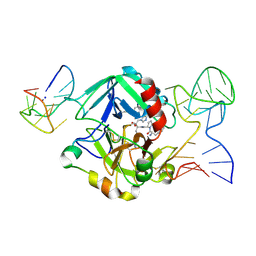 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT3, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
5EW2
 
 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT12, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
5F9T
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, covalent complex with a fluorinated Neu5Ac derivative | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-12-10 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
6GXB
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor JS13 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, ~{N}-[4-[4-[(4-sulfamoylphenyl)carbamoylamino]phenoxy]butyl]ethanamide | | Authors: | Brynda, J, Rezacova, P, Pospisilova, K. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibitor-Polymer Conjugates as a Versatile Tool for Detection and Visualization of Cancer-Associated Carbonic Anhydrase Isoforms
Acs Omega, 4, 2019
|
|
5D8N
 
 | | Tomato leucine aminopeptidase mutant - K354E | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | DuPrez, K.T, Scranton, M.A, Walling, L.L, Fan, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into chaperone-activity enhancement by a K354E mutation in tomato acidic leucine aminopeptidase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FTS
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Tran, F, Westwood, N.J, Naismith, J.H. | | Deposit date: | 2016-01-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric Competitive Inhibitors of the Glucose-1-Phosphate Thymidylyltransferase (Rmla) from Pseudomonas Aeruginosa.
To be Published
|
|
5FKM
 
 | | TetR(D) T103A mutant in complex with anhydrotetracycline and magnesium, I4(1)22 | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Werten, S, Schneider, J, Palm, G.J, Hinrichs, W. | | Deposit date: | 2015-10-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Modular Organisation of Inducer Recognition and Allostery in the Tetracycline Repressor
FEBS J., 283, 2016
|
|
2WQX
 
 | | InlB321_4R: S199R, D200R, G206R, A227R, C242A mutant of the Listeria monocytogenes InlB internalin domain | | Descriptor: | INTERNALIN B | | Authors: | Niemann, H.H, Ferraris, D.M, Heinz, D.W. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand-Mediated Dimerization of the met Receptor Tyrosine Kinase by the Bacterial Invasion Protein Inlb.
J.Mol.Biol., 395, 2010
|
|
5FGY
 
 | |
5FHT
 
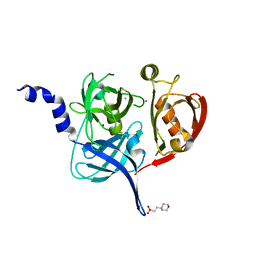 | | HtrA2 protease mutant V226K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Golik, P, Dubin, G, Zurawa-Janicka, D, Lipinska, B, Jarzab, M, Wenta, T, Gieldon, A, Ciarkowski, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Distinct 3D Architecture and Dynamics of the Human HtrA2(Omi) Protease and Its Mutated Variants.
Plos One, 11, 2016
|
|
5DP8
 
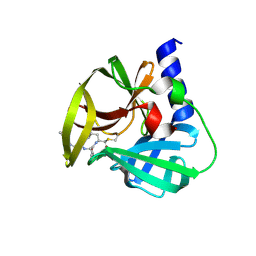 | | Crystal Structure of EV71 3C Proteinase in complex with compound 8 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(2-cyclopropylethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
2Y4M
 
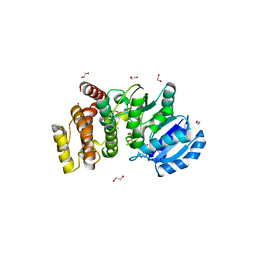 | | MANNOSYLGLYCERATE SYNTHASE IN COMPLEX WITH GDP-Mannose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), ... | | Authors: | Nielsen, M.M, Suits, M.D.L, Yang, M, Barry, C.S, Martinez-Fleites, C, Tailford, L.E, Flint, J.E, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate and Metal Ion Promiscuity in Mannosylglycerate Synthase.
J.Biol.Chem., 286, 2011
|
|
5EEC
 
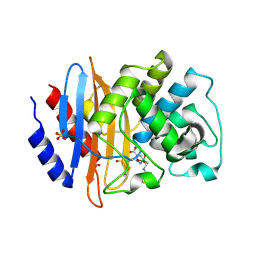 | |
5FBN
 
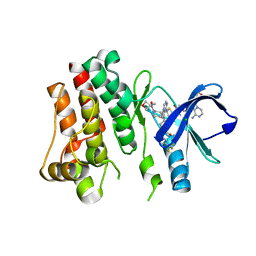 | | BTK kinase domain with inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-[8-azanyl-3-[(3~{R})-1-(3-methyloxetan-3-yl)carbonylpiperidin-3-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | Authors: | Raaijmakers, H.C.A, Vu-Pham, D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 8-Amino-imidazo[1,5-a]pyrazines as Reversible BTK Inhibitors for the Treatment of Rheumatoid Arthritis.
Acs Med.Chem.Lett., 7, 2016
|
|
2VOE
 
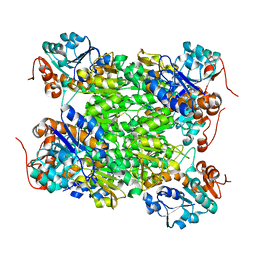 | | Crystal structure of Rv2780 from M. tuberculosis H37Rv | | Descriptor: | ALANINE DEHYDROGENASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Mycobacterium Tuberculosis Secretory Antigen Alanine Dehydrogenase (Rv2780) in Apo and Ternary Complex Forms Captures "Open" and "Closed" Enzyme Conformations.
Proteins, 72, 2008
|
|
6XSY
 
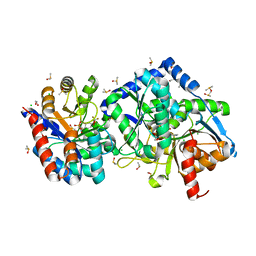 | | The external aldimine crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-S377A with inhibitor 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine (KOU) at the beta-site | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Mueller, L.J, Dunn, M.F. | | Deposit date: | 2020-07-16 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The external aldimine crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-S377A with inhibitor 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine (KOU) at the beta-site.
To be Published
|
|
9AU7
 
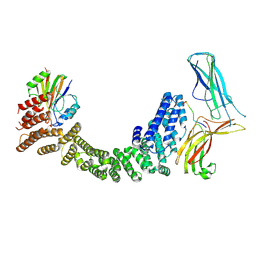 | | Human Retriever VPS35L/VPS29/VPS26C complex bound to SNX17 peptide (Composite Map) | | Descriptor: | Sorting nexin-17, VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 26C, ... | | Authors: | Chen, B, Chen, Z, Han, Y, Boesch, D.J, Juneja, P, Burstein, E, Fung, H.Y.J. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-30 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for Retriever-SNX17 assembly and endosomal sorting.
Nat Commun, 15, 2024
|
|
