4GNC
 
 | | human SMP30/GNL-1,5-AG complex | | Descriptor: | 1,5-anhydro-D-glucitol, CALCIUM ION, Regucalcin | | Authors: | Aizawa, S, Senda, M, Harada, A, Maruyama, N, Ishida, T, Aigaki, T, Ishigami, A, Senda, T. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structural basis of the gamma-lactone-ring formation in ascorbic acid biosynthesis by the senescence marker protein-30/gluconolactonase
Plos One, 8, 2013
|
|
4EP2
 
 | | Crystal Structure of inactive single chain wild-type HIV-1 Protease in Complex with the substrate RT-RH | | Descriptor: | GLYCEROL, PHOSPHATE ION, protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-16 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
6DE7
 
 | |
3GO4
 
 | |
1WJB
 
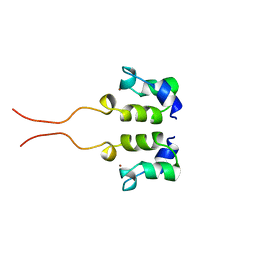 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJF
 
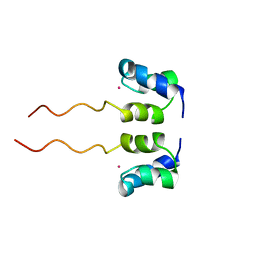 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, 40 STRUCTURES | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
2GSS
 
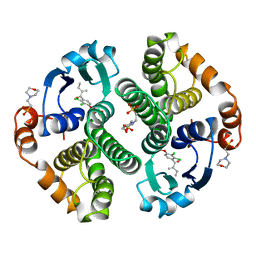 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 IN COMPLEX WITH ETHACRYNIC ACID | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A.J, Rossjohn, J, Parker, M.W. | | Deposit date: | 1996-10-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of the human Pi class glutathione transferase P1-1 in complex with the inhibitor ethacrynic acid and its glutathione conjugate.
Biochemistry, 36, 1997
|
|
3NXP
 
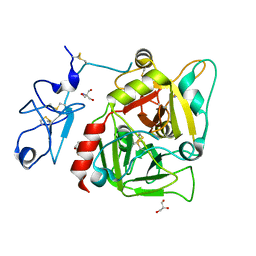 | | Crystal structure of human prethrombin-1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Chen, Z, Bush-Pelc, L.A, Di Cera, E. | | Deposit date: | 2010-07-14 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of prethrombin-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3WXC
 
 | | Crystal Structure of IMP-1 metallo-beta-lactamase complexed with a 3-aminophtalic acid inhibitor | | Descriptor: | 3-(4-hydroxypiperidin-1-yl)benzene-1,2-dicarboxylic acid, Beta-lactamase, ZINC ION | | Authors: | Saito, J, Watanabe, T, Yamada, M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic analysis of IMP-1 metallo-beta-lactamase complexed with a 3-aminophthalic acid derivative, structure-based drug design, and synthesis of 3,6-disubstituted phthalic acid derivative inhibitors
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1B2I
 
 | |
3O3A
 
 | | Human Class I MHC HLA-A2 in complex with the Peptidomimetic ELA-1 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Borbulevych, O.Y, Baker, B.M. | | Deposit date: | 2010-07-23 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of HLA-A*0201 complexed with Melan-A/MART-1(26(27L)-35) peptidomimetics reveal conformational heterogeneity and highlight degeneracy of T cell recognition.
J.Med.Chem., 53, 2010
|
|
3DEI
 
 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | (1S)-2-oxo-1-phenyl-2-[(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-5-yl)amino]ethyl acetate, Caspase-3 | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
3DPH
 
 | | HIV-1 capsid C-terminal domain mutant (L211S) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Igonet, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2008-07-08 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
2YRF
 
 | | Crystal structure of 5-methylthioribose 1-phosphate isomerase from Bacillus subtilis complexed with sulfate ion | | Descriptor: | Methylthioribose-1-phosphate isomerase, SULFATE ION | | Authors: | Tamura, H, Inoue, T, Kai, Y, Matsumura, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: Implications for catalytic mechanism
Protein Sci., 17, 2008
|
|
3DCK
 
 | | X-ray structure of D25N chemical analogue of HIV-1 protease complexed with ketomethylene isostere inhibitor | | Descriptor: | (2S)-2-{[(2R,5S)-5-{[(2S,3S)-2-{[(2S,3R)-2-(acetylamino)-3-hydroxybutanoyl]amino}-3-methylpentanoyl]amino}-2-butyl-4-oxononanoyl]amino}-N~1~-[(2S)-1-amino-5-carbamimidamido-1-oxopentan-2-yl]pentanediamide, Chemical analogue HIV-1 protease | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DS3
 
 | |
3DS4
 
 | |
1OK4
 
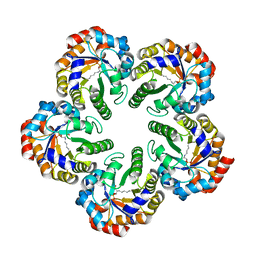 | | Archaeal fructose 1,6-bisphosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-04 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
2WIY
 
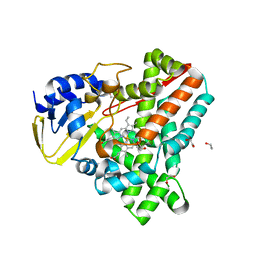 | | Cytochrome P450 XplA heme domain P21212 | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450-LIKE PROTEIN XPLA, IMIDAZOLE, ... | | Authors: | Sabbadin, F, Jackson, R, Bruce, N.C, Grogan, G. | | Deposit date: | 2009-05-18 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The 1.5-A Structure of Xpla-Heme, an Unusual Cytochrome P450 Heme Domain that Catalyzes Reductive Biotransformation of Royal Demolition Explosive.
J.Biol.Chem., 284, 2009
|
|
3TKG
 
 | | crystal structure of HIV model protease precursor/saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | Deposit date: | 2011-08-26 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
3OT3
 
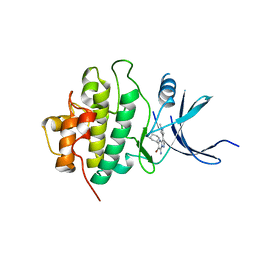 | | X-ray crystal structure of compound 22k bound to human Chk1 kinase domain | | Descriptor: | 5-[(1R,3S)-3-aminocyclohexyl]-6-bromo-3-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidin-7-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-09-10 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Discovery of pyrazolo[1,5-a]pyrimidine-based CHK1 inhibitors: A template-based approach-Part 2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2HOT
 
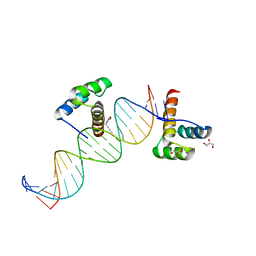 | | Phage selected homeodomain bound to modified DNA | | Descriptor: | 3-PROP-2-YN-1-YL-1,3-OXAZOLIDIN-2-ONE, 5'-D(*AP*TP*CP*CP*GP*GP*GP*GP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*CP*CP*CP*CP*GP*GP*A)-3', ... | | Authors: | Feldman, M.E, Simon, M.D, Shokat, K.M. | | Deposit date: | 2006-07-16 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure and properties of a re-engineered homeodomain protein-DNA interface.
Acs Chem.Biol., 1, 2006
|
|
4H1T
 
 | | X-RAY Structure of the Complex VchUPh with Phosphate ion at 1.92A Resolution. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Sotnichenko, S.E, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-09-11 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | X-RAY Structure of the Complex VchUPh with Phosphate ion at 1.92A Resolution.
TO BE PUBLISHED
|
|
2HQ9
 
 | |
3OOM
 
 | | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507 | | Descriptor: | 1,2-ETHANEDIOL, 1-{3-[6-(tetrahydro-2H-pyran-4-ylamino)imidazo[1,2-b]pyridazin-3-yl]phenyl}ethanone, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Allerston, C, Krojer, T, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507
To be Published
|
|
