7HUL
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0031733-001 (A71EV2A-x3181) | | Descriptor: | 1-methyl-N-[2-(1H-1,2,4-triazol-1-yl)ethyl]-1H-indazole-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
8CGW
 
 | | Insulin-regulated aminopeptidase (IRAP) in complex with an allosteric benzopyran-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Stratikos, E, Giastas, P. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mechanisms of Allosteric Inhibition of Insulin-Regulated Aminopeptidase.
J.Mol.Biol., 436, 2024
|
|
1B6A
 
 | | HUMAN METHIONINE AMINOPEPTIDASE 2 COMPLEXED WITH TNP-470 | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, COBALT (II) ION, METHIONINE AMINOPEPTIDASE | | Authors: | Liu, S, Clardy, J.C. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
5TRJ
 
 | |
3IKD
 
 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-phenylpropyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Matthews, D, Greasley, S, Ferre, R, Parge, H. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6E57
 
 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-B in complex with mixed-linkage heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Koropatkin, N.M, Schnizlein, M, Bahr, C.M.E. | | Deposit date: | 2018-07-19 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
1HAI
 
 | | THE ISOMORPHOUS STRUCTURES OF PRETHROMBIN2, HIRUGEN-AND PPACK-THROMBIN: CHANGES ACCOMPANYING ACTIVATION AND EXOSITE BINDING TO THROMBIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | Tulinsky, A, Vijayalakshmi, J. | | Deposit date: | 1994-06-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin.
Protein Sci., 3, 1994
|
|
5D5M
 
 | | Structure of human MR1-5-OP-RU in complex with human MAIT M33.64 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Keller, A.N, Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2015-08-11 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
3AF2
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with AMPPCP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Pantothenate kinase | | Authors: | Chetnani, B, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2010-02-22 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | M. tuberculosis pantothenate kinase: dual substrate specificity and unusual changes in ligand locations
J.Mol.Biol., 400, 2010
|
|
3PIR
 
 | | Crystal structure of M-RasD41E in complex with GppNHp (type 1) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras | | Authors: | Muraoka, S, Matsumoto, K, Shima, F, Hu, L, Ijiri, Y, Hirai, R, Liao, J, Kataoka, T. | | Deposit date: | 2010-11-07 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of M-RasD41E in complex with GppNHp (type 1)
To be Published
|
|
5QHA
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with PCM-0102951 | | Descriptor: | ACETATE ION, N-[1-(2,3-dihydro-1,4-benzodioxin-6-yl)cyclopentyl]acetamide, Peroxisomal coenzyme A diphosphatase NUDT7 | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-09-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QJ1
 
 | |
7H65
 
 | | THE 1.8 A CRYSTAL STRUCTURE OF HUMAN CHYMASE IN COMPLEX WITH N-[1-[2-[[2-hydroxy-3-methyl-3-(4-methylphenyl)-4-oxocyclobuten-1-yl]-phenylmethyl]-6-methyl-1H-indol-3-yl]-2-methylpropan-2-yl]acetamide | | Descriptor: | Chymase, DIMETHYL SULFOXIDE, N-(1-{2-[(S)-[(3S)-2-hydroxy-3-methyl-3-(4-methylphenyl)-4-oxocyclobut-1-en-1-yl](phenyl)methyl]-6-methyl-1H-indol-3-yl}-2-methylpropan-2-yl)acetamide, ... | | Authors: | Banner, D.W, Benz, J.M, Schlatter, D, Hilpert, H. | | Deposit date: | 2024-04-19 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of human Chymase and Cathepsin G
To be published
|
|
6YLX
 
 | | pre-60S State NE1 (TAP-Flag-Nop53) | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Kater, L, Beckmann, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
4DHF
 
 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2DY8
 
 | |
3AJD
 
 | | Crystal structure of ATRM4 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Putative methyltransferase MJ0026 | | Authors: | Hirano, M, Kuratani, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-01 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of Methanocaldococcus jannaschii Trm4 complexed with sinefungin.
J.Mol.Biol., 401, 2010
|
|
4YEF
 
 | | beta1 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclododextrin | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-1, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-24 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
3IPB
 
 | | Human Transthyretin (TTR) complexed with a palindromic bivalent amyloid inhibitor (11 carbon linker). | | Descriptor: | 2,2'-{undecane-1,11-diylbis[oxy(3,5-dichlorobenzene-4,1-diyl)imino]}dibenzoic acid, Transthyretin | | Authors: | Kolstoe, S.E, Wood, S.P, Pepys, M.B. | | Deposit date: | 2009-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trapping of palindromic ligands within native transthyretin prevents amyloid formation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4QVM
 
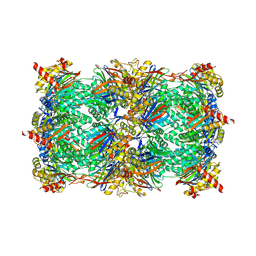 | | yCP beta5-M45A mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5TYZ
 
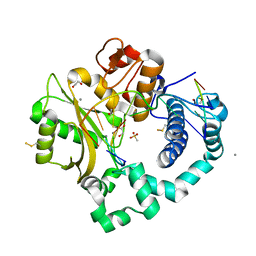 | | DNA Polymerase Mu Product Complex, Mn2+ (960 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
5QRA
 
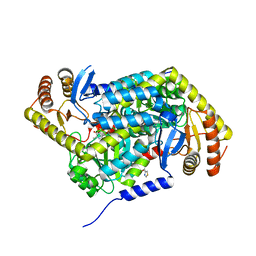 | | PanDDA analysis group deposition -- Crystal Structure of human ALAS2A in complex with Z1101755952 | | Descriptor: | 5-aminolevulinate synthase, erythroid-specific, mitochondrial, ... | | Authors: | Bezerra, G.A, Foster, W, Bailey, H, Shrestha, L, Krojer, T, Talon, R, Brandao-Neto, J, Douangamath, A, Nicola, B.B, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Brennan, P.E, Yue, W.W. | | Deposit date: | 2019-05-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5TR4
 
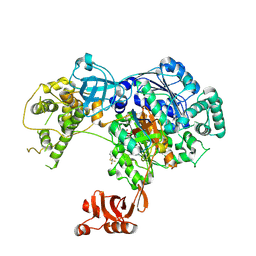 | | Structure of Ubiquitin activating enzyme (Uba1) in complex with ubiquitin and TAK-243 | | Descriptor: | Ubiquitin, Ubiquitin-activating enzyme E1 1, [(1~{R},2~{R},3~{S},4~{R})-2,3-bis(oxidanyl)-4-[[2-[3-(trifluoromethylsulfanyl)phenyl]pyrazolo[1,5-a]pyrimidin-7-yl]amino]cyclopentyl]methyl sulfamate | | Authors: | Sintchak, M.D. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification and Characterization of a Small Molecule Inhibitor of the Ubiquitin Activating Enzyme (TAK-243)
To be published
|
|
7EJH
 
 | | Crystal structure of KRED mutant-F147L/L153Q/Y190P/L199A/M205F/M206F and 2-hydroxyisoindoline-1,3-dione complex | | Descriptor: | 2-oxidanylisoindole-1,3-dione, 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72883928 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
5TZO
 
 | | Computationally Designed Fentanyl Binder - Fen49*-Complex | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase A, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
