3E32
 
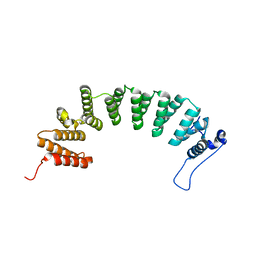 | | Protein farnesyltransferase complexed with FPP and ethylenediamine scaffold inhibitor 2 | | Descriptor: | FARNESYL DIPHOSPHATE, N-benzyl-N-(2-{(4-cyanophenyl)[(1-methyl-1H-imidazol-5-yl)methyl]amino}ethyl)-1-methyl-1H-imidazole-4-sulfonamide, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
7JXF
 
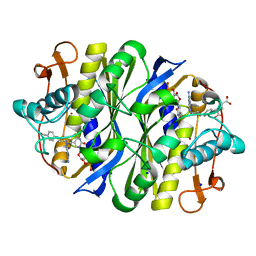 | | E. coli TSase complex with a bi-substrate reaction intermediate diastereomer analog | | Descriptor: | (2S)-2-({4-[({(6S)-2,4-diamino-5-[(1-{(2R,4S,5R)-4-hydroxy-5-[(phosphonooxy)methyl]tetrahydrofuran-2-yl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)methyl]-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl}methyl)amino]benzoyl}amino)pentanedioic acid (non-preferred name), 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE, N-[4-({[(6S)-2,4-diamino-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl]methyl}amino)benzene-1-carbonyl]-L-glutamic acid, ... | | Authors: | Finer-Moore, J, Kholodar, S.A, Stroud, R.M, Kohen, A, Moliner, V, Swiderek, K. | | Deposit date: | 2020-08-27 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caught in Action: X-ray Structure of Thymidylate Synthase with Noncovalent Intermediate Analog.
Biochemistry, 60, 2021
|
|
7JX1
 
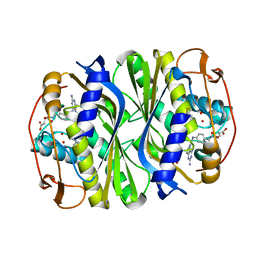 | | E. coli TSase complex with a bi-substrate reaction intermediate analog | | Descriptor: | (2S)-2-({4-[({(6R)-2,4-diamino-5-[(1-{(2R,4S,5R)-4-hydroxy-5-[(phosphonooxy)methyl]tetrahydrofuran-2-yl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)methyl]-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl}methyl)amino]benzoyl}amino)pentanedioic acid (non-preferred name), N-(4-{[(2,4-diamino-7,8-dihydropyrido[3,2-d]pyrimidin-6-yl)methyl]amino}benzene-1-carbonyl)-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Finer-Moore, J, Kholodar, S.A, Stroud, R.M, Kohen, A. | | Deposit date: | 2020-08-26 | | Release date: | 2021-04-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Caught in Action: X-ray Structure of Thymidylate Synthase with Noncovalent Intermediate Analog.
Biochemistry, 60, 2021
|
|
2X2L
 
 | | Crystal Structure of phosphorylated RET tyrosine kinase domain with inhibitor | | Descriptor: | (3Z)-5-AMINO-3-[(4-METHOXYPHENYL)METHYLIDENE]-1,3-DIHYDRO-2H-INDOL-2-ONE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Knowles, P.P, Murray-Rust, J, Kjaer, S, McDonald, N.Q. | | Deposit date: | 2010-01-13 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, structure-activity relationship and crystallographic studies of 3-substituted indolin-2-one RET inhibitors.
Bioorg. Med. Chem., 18, 2010
|
|
5ULM
 
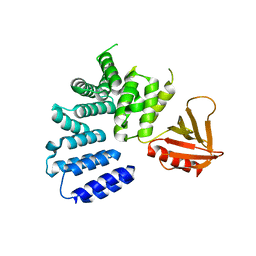 | |
6MTR
 
 | | Crystal structure of VRC43.01 Fab | | Descriptor: | Antibody VRC43.01 Fab heavy chain, Antibody VRC43.01 Fab light chain | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
1OGS
 
 | | human acid-beta-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Dvir, H, Harel, M, McCarthy, A.A, Toker, L, Silman, I, Futerman, A.H, Sussman, J.L. | | Deposit date: | 2003-05-13 | | Release date: | 2003-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structure of Human Acid-Beta-Glucosidase, the Defective Enzyme in Gaucher Disease
Embo Rep., 4, 2003
|
|
6MSC
 
 | | Novel, potent, selective and brain penetrant phosphodiesterase 10A inhibitors | | Descriptor: | 8-fluoro-6-methoxy-3-methyl-1-(3-methylpyridin-4-yl)-3H-pyrazolo[3,4-c]cinnoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Jakob, C.G. | | Deposit date: | 2018-10-16 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Novel, potent, selective, and brain penetrant phosphodiesterase 10A inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
2WPX
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (with AcCoA) | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, ORF14 | | Authors: | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | Deposit date: | 2009-08-11 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
3I4L
 
 | | Structural characterization for the nucleotide binding ability of subunit A with AMP-PNP of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A-TYPE ATP SYNTHASE CATALYTIC SUBUNIT A, ... | | Authors: | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
5V4I
 
 | | Osmium(II)(cymene)(chlorido)2-lysozyme adduct with one binding site | | Descriptor: | Lysozyme C, SODIUM ION, dichloro[(1,2,3,4,5,6-eta)-3-methyl-6-(propan-2-yl)benzene-1,2,4,5-tetrayl]osmium | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The metalation of hen egg white lysozyme impacts protein stability as shown by ion mobility mass spectrometry, differential scanning calorimetry, and X-ray crystallography.
Chem. Commun. (Camb.), 53, 2017
|
|
4RLT
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with Fisetin | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 3,7,3',4'-TETRAHYDROXYFLAVONE, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
6D9F
 
 | | Protein 60 with aldehyde deformylating oxidase activity from Kitasatospora setae | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Putative VlmB homolog, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2018-04-28 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Biochemistry, 59, 2020
|
|
3RM6
 
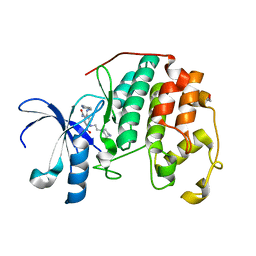 | | CDK2 in complex with inhibitor KVR-2-80 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-5-nitro-4-{[2-(piperazin-1-yl)ethyl]amino}benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
4GAF
 
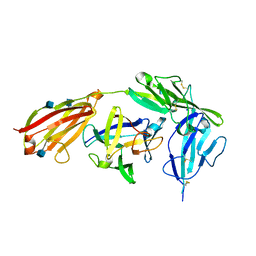 | | Crystal structure of EBI-005, a chimera of human IL-1beta and IL-1Ra, bound to human Interleukin-1 receptor type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EBI-005, Interleukin-1 receptor type 1, ... | | Authors: | Hou, J, Townson, S.A, Kovalchin, J.T, Masci, A, Kiner, O, Shu, Y, King, B, Thomas, C, Garcia, K.C, Furfine, E.S, Barnes, T.M. | | Deposit date: | 2012-07-25 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design of a superior cytokine antagonist for topical ophthalmic use.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5AOL
 
 | |
4GCJ
 
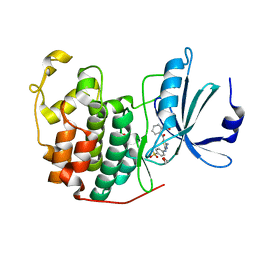 | | CDK2 in complex with inhibitor RC-3-89 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(2-nitrobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2012-07-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3HXZ
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS G237A in complex with AlaSA | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
2X1E
 
 | | The crystal structure of mature acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum in complex 6- aminopenicillanic acid | | Descriptor: | (2S,5R,6R)-6-AMINO-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ACYL-COENZYME, CHLORIDE ION, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
3EHS
 
 | |
5TM1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,5-bis(2-fluoro-4-hydroxyphenyl)thiophene 1-oxide | | Descriptor: | 2,5-bis(2-fluoro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, (E)-3-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)acrylic acid | | Descriptor: | 3-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}prop-2-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5W3P
 
 | | ANTIBODY C706 IN COMPLEX WTH BETA-AMYLOID PEPTIDE 1-16 | | Descriptor: | Amyloid beta A4 protein, Antibody C706 Fab HEAVY CHAIN, Antibody C706 Fab LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2017-06-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A coiled conformation of amyloid-beta recognized by antibody C706.
Alzheimers Res Ther, 9, 2017
|
|
7KI1
 
 | | Taspoglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs Protein | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, X, Belousoff, M.J, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2020-10-22 | | Release date: | 2021-08-04 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and dynamics of semaglutide- and taspoglutide-bound GLP-1R-Gs complexes.
Cell Rep, 36, 2021
|
|
3E30
 
 | | Protein farnesyltransferase complexed with FPP and ethylene diamine inhibitor 4 | | Descriptor: | FARNESYL DIPHOSPHATE, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
