6TT4
 
 | |
3P30
 
 | | crystal structure of the cluster II Fab 1281 in complex with HIV-1 gp41 ectodomain | | Descriptor: | 1281 Fab heavy chain, 1281 Fab light chain, HIV-1 gp41 | | Authors: | Frey, G, Chen, J, Rits-Volloch, S, Freeman, M.M, Zolla-Pazner, S, Chen, B. | | Deposit date: | 2010-10-04 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies.
Nat.Struct.Mol.Biol., 17, 2010
|
|
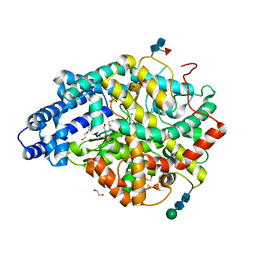
1LLF
 
 | | Cholesterol Esterase (Candida Cylindracea) Crystal Structure at 1.4A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase 3, TRICOSANOIC ACID | | Authors: | Pletnev, V, Addlagatta, A, Wawrzak, Z, Duax, W. | | Deposit date: | 2002-04-28 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Three-dimensional structure of homodimeric cholesterol esterase-ligand complex at 1.4 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
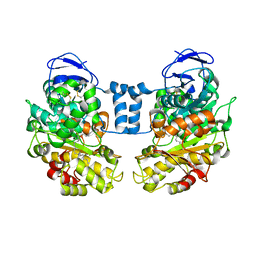
3P8G
 
 | | Crystal Structure of MT-SP1 in complex with benzamidine | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, GLUTATHIONE, ... | | Authors: | Yuan, C, Huang, M, Chen, L. | | Deposit date: | 2010-10-13 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of catalytic domain of Matriptase in complex with Sunflower trypsin inhibitor-1.
Bmc Struct.Biol., 11, 2011
|
|
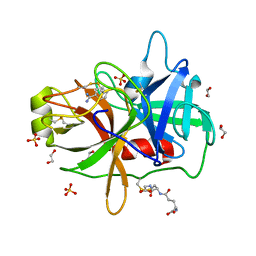
1OM1
 
 | | Crystal structure of maize CK2 alpha in complex with IQA | | Descriptor: | (5-OXO-5,6-DIHYDRO-INDOLO[1,2-A]QUINAZOLIN-7-YL)-ACETIC ACID, Casein kinase II, alpha chain | | Authors: | Battistutta, R, De Moliner, E, Zanotti, G. | | Deposit date: | 2003-02-24 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Biochemical and three-dimensional-structural study of the specific inhibition of protein kinase CK2 by [5-oxo-5,6-dihydroindolo-(1,2-a)quinazolin-7-yl]acetic acid (IQA).
Biochem.J., 374, 2003
|
|
4LOY
 
 | | Crystal Structure Analysis of thrombin in complex with compound D57, 5-Chlorothiophene-2-carboxylic acid [(S)-2-[2-methyl-3-(2- oxopyrrolidin-1-yl)benzenesulfonylamino]-3-(4-methylpiperazin-1- yl)-3-oxopropyl]amide (SAR107375) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-Chloro-thiophene-2-carboxylic acid [(S)-2-[2-chloro-5-fluoro-3-(2-oxo-piperidin-1-yl)-benzenesulfonylamino]-3-(4-methyl-piperazin-1-yl)-3-oxo-propyl]-amide, Hirudin variant-2, ... | | Authors: | Stehlin-Gaon, C, Bocskei, Z. | | Deposit date: | 2013-07-14 | | Release date: | 2014-06-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | 5-Chlorothiophene-2-carboxylic acid [(S)-2-[2-methyl-3-(2-oxopyrrolidin-1-yl)benzenesulfonylamino]-3-(4-methylpiperazin-1-yl)-3-oxopropyl]amide (SAR107375), a selective and potent orally active dual thrombin and factor Xa inhibitor.
J.Med.Chem., 56, 2013
|
|
5UFZ
 
 | | HIV-1 Protease complexed with Inhibitor: (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2S)-1-(1-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}cyclopropyl)-1-hydroxy-3-phenylpropan-2-yl]carbamate | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2S)-1-(1-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}cyclopropyl)-1-hydroxy-3-phenylpropan-2-yl]carbamate, Protease | | Authors: | Zachary, N.E.R. | | Deposit date: | 2017-01-06 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Synthesis of Novel HIV-1 Protease Inhibitors via Diasteroselective Henry Reaction with Nitrocyclopropane
TO BE PUBLISHED
|
|
4DOQ
 
 | | Crystal structure of the complex of Porcine Pancreatic Trypsin with 1/2SLPI | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Antileukoproteinase, CALCIUM ION, ... | | Authors: | Fukushima, K, Takimoto-Kamimura, M. | | Deposit date: | 2012-02-10 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure basis 1/2SLPI and porcine pancreas trypsin interaction
J.SYNCHROTRON RADIAT., 20, 2013
|
|
1NLR
 
 | | ENDO-1,4-BETA-GLUCANASE CELB2, CELLULASE, NATIVE STRUCTURE | | Descriptor: | ENDO-1,4-BETA-GLUCANASE | | Authors: | Sulzenbacher, G, Dupont, C, Davies, G.J. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 A resolution.
Biochemistry, 36, 1997
|
|
3DYA
 
 | | HIV-1 RT with non-nucleoside inhibitor annulated Pyrazole 1 | | Descriptor: | 3-[6-bromo-2-fluoro-3-(1H-pyrazolo[3,4-c]pyridazin-3-ylmethyl)phenoxy]-5-chlorobenzonitrile, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, p51 RT | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of annulated pyrazoles as inhibitors of HIV-1 reverse transcriptase
J.Med.Chem., 51, 2008
|
|
1NPA
 
 | | crystal structure of HIV-1 protease-hup | | Descriptor: | (3S)-TETRAHYDROFURAN-3-YL (1R,2S)-3-[4-((1R)-2-{[(S)-AMINO(HYDROXY)METHYL]OXY}-2,3-DIHYDRO-1H-INDEN-1-YL)-2-BENZYL-3-OXOPYRROLIDIN-2-YL]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, POL polyprotein | | Authors: | Smith III, A.B, Hirschmann, R, Pasternak, A, Yao, W, Sprengeler, P.A, Holloway, M.K, Kuo, L.C, Chen, Z, Darke, P.L, Schleif, W.A. | | Deposit date: | 2003-01-17 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable pyrrolinone inhibitor of HIV-1 protease: computational analysis and X-ray crystal structure of the enzyme complex.
J.MED.CHEM., 40, 1997
|
|
6B2E
 
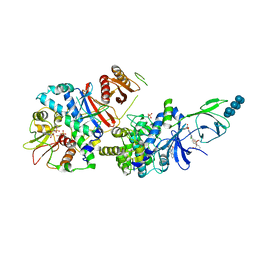 | | Structure of full length human AMPK (a2b2g1) in complex with a small molecule activator SC4. | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
3FD0
 
 | |
1NWG
 
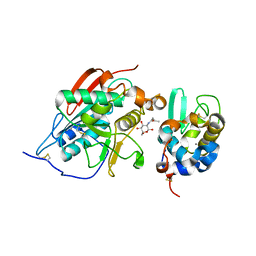 | | BETA-1,4-GALACTOSYLTRANSFERASE COMPLEX WITH ALPHA-LACTALBUMIN AND N-BUTANOYL-GLUCOAMINE | | Descriptor: | 2-(butanoylamino)-2-deoxy-beta-D-glucopyranose, Alpha-lactalbumin, CALCIUM ION, ... | | Authors: | Ramakrishnan, B, Shah, P.S, Qasba, P.K. | | Deposit date: | 2003-02-06 | | Release date: | 2003-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | ALPHA-LACTALBUMIN (LA) STIMULATES MILK
BETA-1,4-GALACTOSYLTRANSFERASE I (BETA 4GAL-T1) TO
TRANSFER GLUCOSE FROM UDP-GLUCOSE TO
N-ACETYLGLUCOSAMINE. CRYSTAL STRUCTURE OF BETA
4GAL-T1 X LA COMPLEX WITH UDP-GLC.
J.Biol.Chem., 276, 2001
|
|
4EUU
 
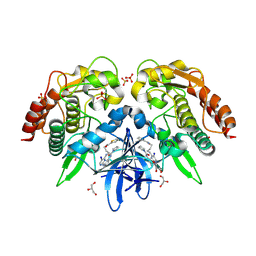 | | Structure of BX-795 Complexed with Human TBK1 Kinase Domain Phosphorylated on Ser172 | | Descriptor: | GLYCEROL, IODIDE ION, N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, ... | | Authors: | Ma, X, Helgason, E, Phung, Q.T, Quan, C.L, Iyer, R.S, Lee, M.W, Bowman, K.K, Starovasnik, M.A, Dueber, E.C. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3SWQ
 
 | | E. Cloacae MurA in complex with Enolpyruvyl-UNAG | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhu, J.-Y, Yang, Y, Schonbrunn, E. | | Deposit date: | 2011-07-14 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
3GF6
 
 | |
3G0J
 
 | | Crystal Structure of the fifth Bromodomain of Human Poly-bromodomain containing protein 1 (PB1) | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Keates, T, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the fifth Bromodomain of Human Poly-bromodomain containing protein 1 (PB1)
To be Published
|
|
1NPW
 
 | | Crystal structure of HIV protease complexed with LGZ479 | | Descriptor: | CARBAMIC ACID 1-{5-BENZYL-5-[2-HYDROXY-4-PHENYL-3-(TETRAHYDRO-FURAN- 3-YLOXYCARBONYLAMINO)-BUTYL]-4-OXO-4,5-DIHYDRO-1H-PYRROL-3-YL}- INDAN-2-YL ESTER, POL polyprotein | | Authors: | Smith III, A.B. | | Deposit date: | 2003-01-20 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors.
J.Med.Chem., 46, 2003
|
|
5OE3
 
 | | Crystal structure of the N-terminal domain of PqsA in complex with anthraniloyl-AMP (crystal form 1) | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-[(2-aminobenzoyl)oxy](hydroxy)phosphoryl]adenosine, ACETATE ION, ... | | Authors: | Witzgall, F, Ewert, W, Blankenfeldt, W. | | Deposit date: | 2017-07-07 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structures of the N-Terminal Domain of PqsA in Complex with Anthraniloyl- and 6-Fluoroanthraniloyl-AMP: Substrate Activation in Pseudomonas Quinolone Signal (PQS) Biosynthesis.
Chembiochem, 18, 2017
|
|
6B3W
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit,Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
4QRC
 
 | | Crystal Structure of the Tyrosine Kinase Domain of FGF Receptor 4 in Complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | DFG-out Mode of Inhibition by an Irreversible Type-1 Inhibitor Capable of Overcoming Gate-Keeper Mutations in FGF Receptors.
Acs Chem.Biol., 10, 2015
|
|
4QTO
 
 | | 1.65 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-modified Cys289 and PEG molecule in active site | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R4H
 
 | | Crystal structure of non-neutralizing, A32-like antibody 2.2c in complex with HIV-1 Env gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c, Heavy chain, ... | | Authors: | Mclellan, J, Acharya, P, Huang, C.-C, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.28 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
1GUH
 
 | | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the MU and PI class enzymes | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1-1, S-BENZYL-GLUTATHIONE | | Authors: | Sinning, I, Kleywegt, G.J, Jones, T.A. | | Deposit date: | 1993-02-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure determination and refinement of human alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes.
J.Mol.Biol., 232, 1993
|
|
