5PH2
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D in complex with N09597a | | Descriptor: | 1,2-ETHANEDIOL, 6-methylpyridine-3-carboxamide, Lysine-specific demethylase 4D, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1I8D
 
 | | CRYSTAL STRUCTURE OF RIBOFLAVIN SYNTHASE | | Descriptor: | RIBOFLAVIN SYNTHASE | | Authors: | Liao, D.-I, Wawrzak, Z, Calabrese, J.C, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-03-13 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of riboflavin synthase.
Structure, 9, 2001
|
|
7K96
 
 | |
4ZZX
 
 | | Structure of PARP2 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-(3-methoxypropyl)-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, POLY [ADP-RIBOSE] POLYMERASE 2 | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
3G36
 
 | | Crystal structure of the human DPY-30-like C-terminal domain | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Wang, X, Lou, Z, Bartlam, M, Rao, Z. | | Deposit date: | 2009-02-02 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the C-terminal domain of human DPY-30-like protein: A component of the histone methyltransferase complex
J.Mol.Biol., 390, 2009
|
|
6F8R
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-54 | | Descriptor: | (2~{S})-1-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
7KDZ
 
 | |
2W5U
 
 | | Flavodoxin from Helicobacter pylori in complex with the C3 inhibitor | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, [2-(5-amino-4-cyano-1H-pyrazol-1-yl)-5-(trifluoromethyl)phenyl](hydroxy)oxoammonium | | Authors: | Cremades, N, Perez-Dorado, I, Hermoso, J.A, Martinez-Julvez, M, Sancho, J. | | Deposit date: | 2008-12-12 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of Specific Flavodoxin Inhibitors as Potential Therapeutic Agents Against Helicobacter Pylori Infection.
Acs Chem.Biol., 4, 2009
|
|
3W27
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
1Q5K
 
 | | crystal structure of Glycogen synthase kinase 3 in complexed with inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-(4-METHOXYBENZYL)-N'-(5-NITRO-1,3-THIAZOL-2-YL)UREA | | Authors: | Bhat, R, Xue, Y, Berg, S, Hellberg, S, Ormo, M, Nilsson, Y, Radesater, A.C, Jerning, E, Markgren, P.O, Borgegard, T, Nylof, M, Gimenez-Cassina, A, Hernandez, F, Lucas, J.J, Diaz-Mido, J, Avila, J. | | Deposit date: | 2003-08-08 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418.
J.Biol.Chem., 278, 2003
|
|
5U4K
 
 | | NMR structure of the complex between the KIX domain of CBP and the transactivation domain 1 of p65 | | Descriptor: | CREB-binding protein, Transcription factor p65 | | Authors: | Lecoq, L, Raiola, L, Chabot, P.R, Cyr, N, Arseneault, G, Omichinski, J.G. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
6AOK
 
 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal TEV protease cleavage sequence | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ceg4, ... | | Authors: | Stogios, P.J, Cuff, M.E, Nocek, B, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | Deposit date: | 2017-08-16 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
3G7W
 
 | | Islet Amyloid Polypeptide (IAPP or Amylin) Residues 1 to 22 fused to Maltose Binding Protein | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | Authors: | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
3VUS
 
 | | Escherichia coli PgaB N-terminal domain | | Descriptor: | ACETATE ION, MERCURY (II) ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | Authors: | Nishiyama, T, Noguchi, H, Yoshida, H, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-07-05 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: a circularly permuted member of the carbohydrate esterase 4 family
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5E2K
 
 | | Crystal structure of human carbonic anhydrase II in complex with the 4-(3-aminophenyl)benzenesulfonamide inhibitor | | Descriptor: | 3'-aminobiphenyl-4-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T. | | Deposit date: | 2015-10-01 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 4-Arylbenzenesulfonamides as Human Carbonic Anhydrase Inhibitors (hCAIs): Synthesis by Pd Nanocatalyst-Mediated Suzuki-Miyaura Reaction, Enzyme Inhibition, and X-ray Crystallographic Studies.
J.Med.Chem., 59, 2016
|
|
2VQT
 
 | | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases | | Descriptor: | (2Z,3R,4S,5R,6R)-2-[(2-aminoethyl)imino]-6-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, BETA-MANNOSIDASE, ... | | Authors: | Tailford, L.E, Offen, W.A, Smith, N.L, Dumon, C, Moreland, C, Gratien, J, Heck, M.P, Stick, R.V, Bleriot, Y, Vasella, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Evidence for a Boat-Like Transition State in Beta-Mannosidases.
Nat.Chem.Biol., 4, 2008
|
|
3ZKJ
 
 | | Crystal Structure of Ankyrin Repeat and Socs Box-Containing Protein 9 (Asb9) in Complex with Elonginb and Elonginc | | Descriptor: | 1,2-ETHANEDIOL, ANKYRIN REPEAT AND SOCS BOX PROTEIN 9, CHLORIDE ION, ... | | Authors: | Muniz, J.R.C, Guo, K, Zhang, Y, Ayinampudi, V, Savitsky, P, Keates, T, Filippakopoulos, P, Vollmar, M, Yue, W.W, Krojer, T, Ugochukwu, E, von Delft, F, Knapp, S, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2013-01-23 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
4EFO
 
 | | Crystal structure of the ubiquitin-like domain of human TBK1 | | Descriptor: | Serine/threonine-protein kinase TBK1 | | Authors: | Li, J, Li, J, Miyahira, A, Sun, J, Liu, Y, Cheng, G, Liang, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Crystal structure of the ubiquitin-like domain of human TBK1.
Protein Cell, 3, 2012
|
|
5GHV
 
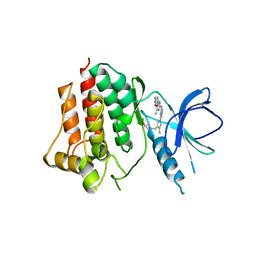 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-({1-[2-({3,5-dimethyl-4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]-4-methyl-1H-pyrrol-3-yl}methyl)azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
5AFG
 
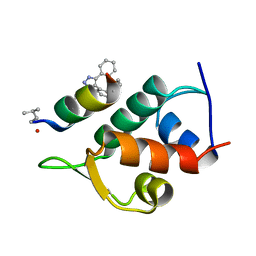 | | Structure of the Stapled Peptide Bound to Mdm2 | | Descriptor: | 1,8-DIETHYL-1,8-DIHYDRODIBENZO[3,4:7,8][1,2,3]TRIAZOLO[4',5':5,6]CYCLOOCTA[1,2-D][1,2,3]TRIAZOLE, E3 UBIQUITIN-PROTEIN LIGASE MDM2, STAPLED PEPTIDE | | Authors: | Lau, Y.H, Wu, Y, Rossmann, M, de Andrade, P, Tan, Y.S, McKenzie, G.J, Venkitaraman, A.R, Hyvonen, M, Spring, D.R. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Double Strain-Promoted Macrocyclization for the Rapid Selection of Cell-Active Stapled Peptides.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
9E8P
 
 | | Crystal Structure of GABARAP-ATG3 conjugate | | Descriptor: | 1,2-ETHANEDIOL, Gamma-aminobutyric acid receptor-associated protein, SULFATE ION, ... | | Authors: | Ohashi, K, Otomo, T. | | Deposit date: | 2024-11-05 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the GABARAP-ATG3 Backside Interaction and Apo ATG3 Conformation.
Biochemistry, 64, 2025
|
|
6B9I
 
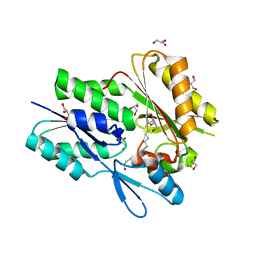 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with 14-Methylhexadecanoic Acid (Anteiso C17:0) to 1.93 Angstrom resolution | | Descriptor: | (14S)-14-methylhexadecanoic acid, Fatty acid Kinase (Fak) B1, GLYCEROL, ... | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-10-10 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Acyl-chain selectivity and physiological roles ofStaphylococcus aureusfatty acid-binding proteins.
J. Biol. Chem., 294, 2019
|
|
4EHG
 
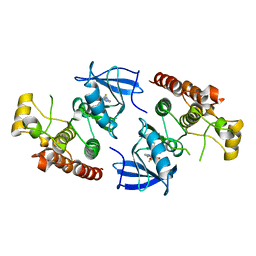 | | B-Raf Kinase Domain in Complex with an Aminopyridimine-based Inhibitor | | Descriptor: | N-{2,4-difluoro-3-[({6-[(2-hydroxyethyl)amino]pyrimidin-4-yl}carbamoyl)amino]phenyl}propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C. | | Deposit date: | 2012-04-02 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent and selective aminopyrimidine-based B-raf inhibitors with favorable physicochemical and pharmacokinetic properties.
J.Med.Chem., 55, 2012
|
|
3ZZF
 
 | | Crystal structure of the amino acid kinase domain from Saccharomyces cerevisiae acetylglutamate kinase complexed with its substrate N- acetylglutamate | | Descriptor: | 1,2-ETHANEDIOL, ACETYLGLUTAMATE KINASE, CHLORIDE ION, ... | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-09-01 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
4QBK
 
 | | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with amino acyl-tRNA analogue at 1.77 Angstrom resolution | | Descriptor: | 3'-deoxy-3'-[(O-methyl-L-tyrosyl)amino]adenosine, GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
