4JR0
 
 | | Human procaspase-3 bound to Ac-DEVD-CMK | | Descriptor: | Ac-DEVD-CMK, CHLORIDE ION, Procaspase-3 | | Authors: | Thomsen, N.D, Wells, J.A. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JRH
 
 | | Crystal structure of beta-ketoacyl-ACP synthase II (FabF) from Vibrio Cholerae (space group P43) at 2.2 Angstrom | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, ACETATE ION | | Authors: | Hou, J, Chruszcz, M, Shabalin, I.G, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: |
|
|
4JLH
 
 | |
4JUT
 
 | | Crystal structure of a mutant fragment of Human HSPB6 | | Descriptor: | GLYCEROL, Heat shock protein beta-6 | | Authors: | Weeks, S.D, Baranova, E.V, Beelen, S, Heirbaut, M, Gusev, N.B, Strelkov, S.V. | | Deposit date: | 2013-03-25 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Molecular structure and dynamics of the dimeric human small heat shock protein HSPB6.
J.Struct.Biol., 185, 2014
|
|
4JWG
 
 | | Crystal structure of spTrm10(74) | | Descriptor: | ACETIC ACID, tRNA (guanine(9)-N1)-methyltransferase | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2013-03-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
4JQU
 
 | | Crystal structure of Ubc7p in complex with the U7BR of Cue1p | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Coupling of ubiquitin conjugation to ER degradation protein 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liang, Y.-H, Metzger, M.B, Weissman, A.M, Ji, X. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A Structurally Unique E2-Binding Domain Activates Ubiquitination by the ERAD E2, Ubc7p, through Multiple Mechanisms.
Mol.Cell, 50, 2013
|
|
4JRC
 
 | | Distal Stem I region from G. kaustophilus glyQS T box RNA | | Descriptor: | Distal Stem I region of the glyQS T box leader RNA, MAGNESIUM ION | | Authors: | Grigg, J.C, Ke, A. | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.673 Å) | | Cite: | T box RNA decodes both the information content and geometry of tRNA to affect gene expression.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KAL
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinoline-3-carboxylic acid | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinoline-3-carboxylic acid
To be Published
|
|
4KAG
 
 | | Crystal structure analysis of a single amino acid deletion mutation in EGFP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein, ... | | Authors: | Arpino, J.A.J, Rizkallah, P.J. | | Deposit date: | 2013-04-22 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural and dynamic changes associated with beneficial engineered single-amino-acid deletion mutations in enhanced green fluorescent protein.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KCE
 
 | |
4KG4
 
 | |
4KK4
 
 | |
4KHR
 
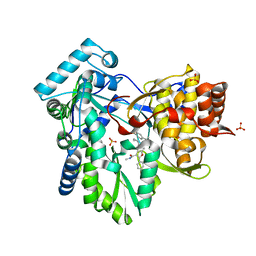 | | HCV NS5B GT1A C316Y with GSK5852 | | Descriptor: | NS5B RNA-dependent RNA polymerase, SULFATE ION, [4-({[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl](methylsulfonyl)amino}methyl)-2-fluorophenyl]boronic acid | | Authors: | Williams, S.P, Kahler, K.M, Shotwell, J.B. | | Deposit date: | 2013-05-01 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of a Potent Boronic Acid Derived Inhibitor of the HCV RNA-Dependent RNA Polymerase.
J.Med.Chem., 57, 2014
|
|
4KMA
 
 | |
4KK7
 
 | |
4KNI
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-Chloro-4-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2013-05-10 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Benzenesulfonamides with pyrimidine moiety as inhibitors of human carbonic anhydrases I, II, VI, VII, XII, and XIII
Bioorg.Med.Chem., 21, 2013
|
|
4WIP
 
 | | DIX domain of human Dvl2 | | Descriptor: | PENTAETHYLENE GLYCOL, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Fiedler, M, Bienz, M, Madrzak, J, Chin, J.W. | | Deposit date: | 2014-09-26 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Ubiquitination of the Dishevelled DIX domain blocks its head-to-tail polymerization.
Nat Commun, 6, 2015
|
|
4KNC
 
 | | Structural and functional characterization of Pseudomonas aeruginosa AlgX | | Descriptor: | Alginate biosynthesis protein AlgX | | Authors: | Riley, L.M, Weadge, J.T, Baker, P, Robinson, H, Codee, J.D.C, Tipton, P.A, Ohman, D.E, Howell, P.L. | | Deposit date: | 2013-05-09 | | Release date: | 2013-06-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural and Functional Characterization of Pseudomonas aeruginosa AlgX: ROLE OF AlgX IN ALGINATE ACETYLATION.
J.Biol.Chem., 288, 2013
|
|
4KQF
 
 | | Crystal structure of CobT E174A complexed with adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
4KR8
 
 | |
4KRP
 
 | |
4K1F
 
 | | Crystal structure of reduced tryparedoxin peroxidase from leishmania major at 2.34 A resolution | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ilari, A, Fiorillo, A, Di Chiaro, F. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based discovery of the first non-covalent inhibitors of Leishmania major tryparedoxin peroxidase by high throughput docking
Sci Rep, 5, 2015
|
|
4K2M
 
 | | Crystal structure of ntda from bacillus subtilis in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, ACETATE ION, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2013-04-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
4K2X
 
 | |
4E28
 
 | | Structure of human thymidylate synthase in inactive conformation with a novel non-peptidic inhibitor | | Descriptor: | 2-{(2Z,5S)-4-hydroxy-2-[(2E)-(2-hydroxybenzylidene)hydrazinylidene]-2,5-dihydro-1,3-thiazol-5-yl}-N-[3-(trifluoromethyl)phenyl]acetamide, 2-{(5S)-2-[(2E)-2-(2-hydroxybenzylidene)hydrazinyl]-4-oxo-4,5-dihydro-1,3-thiazol-5-yl}-N-[3-(trifluoromethyl)phenyl]acetamide, SULFATE ION, ... | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M, Costi, M.P. | | Deposit date: | 2012-03-07 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Inhibitor of ovarian cancer cells growth by virtual screening: a new thiazole derivative targeting human thymidylate synthase.
J.Med.Chem., 55, 2012
|
|
