3X2Q
 
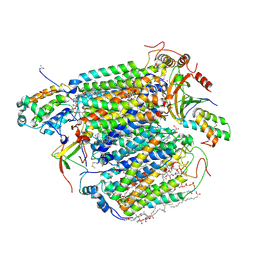 | | X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2014-12-26 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
6R3G
 
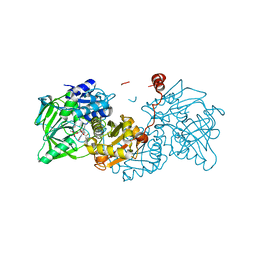 | |
8TBH
 
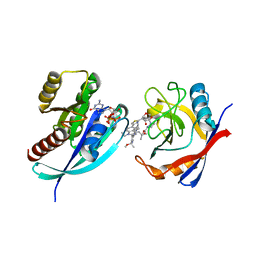 | | Tricomplex of RMC-7977, KRAS G12R, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Bar Ziv, T, Zhang, D, Tomlinson, A.C.A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 629, 2024
|
|
6RQP
 
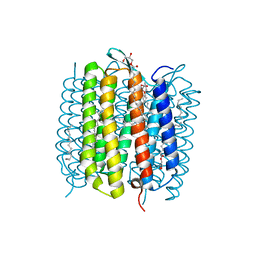 | | Steady-state-SMX dark state structure of bacteriorhodopsin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
3T45
 
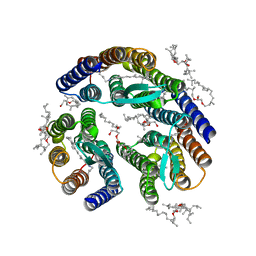 | | Crystal structure of bacteriorhodopsin mutant A215T, a phototaxis signaling mutant at 3.0 A resolution | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin (GROUND STATE), RETINAL | | Authors: | Ozorowski, G, Luecke, H. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A transporter converted into a sensor, a phototaxis signaling mutant of bacteriorhodopsin at 3.0 angstrom.
J.Mol.Biol., 415, 2012
|
|
6S7O
 
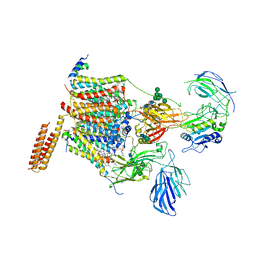 | | Cryo-EM structure of human oligosaccharyltransferase complex OST-A | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R},3~{R},4~{R},5'~{S},6~{S},7~{R},8~{S},9~{R},12~{R},13~{R},15~{S},16~{S},18~{R})-5',7,9,13-tetramethyl-3,15-bis(oxidanyl)spiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-16-yl]oxy-oxane-3,4,5-triol, (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit, ... | | Authors: | Ramirez, A.S, Kowal, J, Locher, K.P. | | Deposit date: | 2019-07-05 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B.
Science, 366, 2019
|
|
1VF5
 
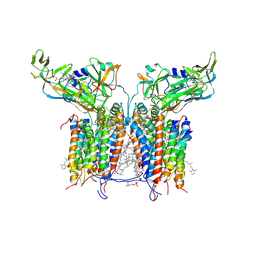 | | Crystal Structure of Cytochrome b6f Complex from M.laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | Authors: | Kurisu, G, Zhang, H, Smith, J.L, Cramer, W.A. | | Deposit date: | 2004-04-08 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Cytochrome B6F Complex of Oxygenic Photosynthesis: Tuning the Cavity
Science, 302, 2003
|
|
6SES
 
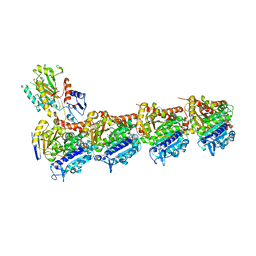 | | Tubulin-B2 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Guo, B, Rodriguez-Gabin, A, Prota, A.E, Muehlethaler, T, Zhang, N, Ye, K, Steinmetz, M.O, Band Horwitz, S, Smith III, A.B, McDaid, H.M. | | Deposit date: | 2019-07-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Refinement of the Tubulin Ligand (+)-Discodermolide to Attenuate Chemotherapy-Mediated Senescence.
Mol.Pharmacol., 98, 2020
|
|
6R3F
 
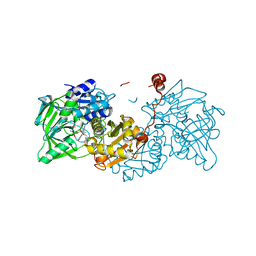 | |
1W5C
 
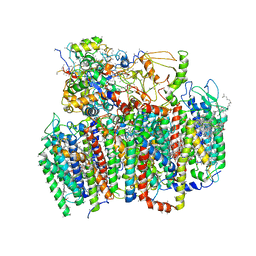 | | Photosystem II from Thermosynechococcus elongatus | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Biesiadka, J, Loll, B, Kern, J, Irrgang, K.-D, Saenger, W. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Cyanobacterial Photosystem II at 3.2 A Resolution: A Closer Look at the Mn- Cluster
Phys.Chem.Chem.Phys., 6, 2004
|
|
3SE2
 
 | | Human poly(ADP-ribose) polymerase 14 (PARP14/ARTD8) - catalytic domain in complex with 6(5H)-phenanthridinone | | Descriptor: | 3-aminobenzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Graslund, S, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Thorsell, A.G, Tresaugues, L, Weigelt, J, Siponen, M.I, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
1YLZ
 
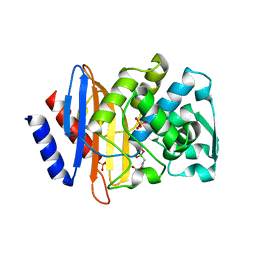 | | X-ray crystallographic structure of CTX-M-14 beta-lactamase complexed with ceftazidime-like boronic acid | | Descriptor: | BETA-LACTAMASE CTX-M-14, PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Chen, Y, Shoichet, B, Bonnet, R. | | Deposit date: | 2005-01-19 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure, Function, and Inhibition along the Reaction Coordinate of CTX-M beta-Lactamases.
J.Am.Chem.Soc., 127, 2005
|
|
1WOU
 
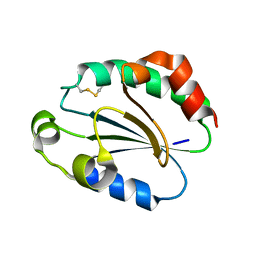 | | Crystal Structure of human Trp14 | | Descriptor: | thioredoxin -related protein, 14 kDa | | Authors: | Woo, J.R, Kim, S.J, Jeong, W, Cho, Y.H, Lee, S.C, Chung, Y.J, Rhee, S.G, Ryu, S.E. | | Deposit date: | 2004-08-25 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of cellular redox regulation by human TRP14
J.Biol.Chem., 279, 2004
|
|
3SMI
 
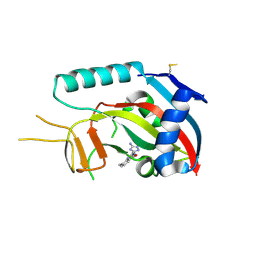 | | Human poly(ADP-ribose) polymerase 14 (Parp14/Artd8) - catalytic domain in complex with a quinazoline inhibitor | | Descriptor: | 2-{[(3-amino-1H-1,2,4-triazol-5-yl)sulfanyl]methyl}-8-methylquinazolin-4(3H)-one, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Graslund, S, Kouznetsova, E, Nordlund, P, Nyman, T, Thorsell, A.G, Tresaugues, L, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
2H2I
 
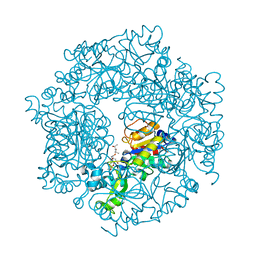 | | The Structural basis of Sirtuin Substrate Affinity | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, NAD-dependent deacetylase, ZINC ION | | Authors: | Cosgrove, M.S, Wolberger, C. | | Deposit date: | 2006-05-18 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of sirtuin substrate affinity
Biochemistry, 45, 2006
|
|
6GTU
 
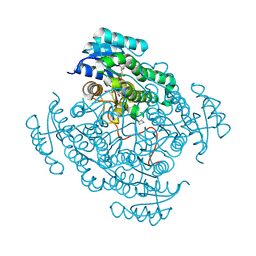 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with fragment J6 | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, CHLORIDE ION, N-(1,3-benzodioxol-5-ylmethyl)cyclopentanamine, ... | | Authors: | Bertoletti, N, Heine, A, Marchais-Oberwinkler, S, Klebe, G. | | Deposit date: | 2018-06-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray Crystallographic Fragment screening and Hit Optimization
To Be Published
|
|
7ZKO
 
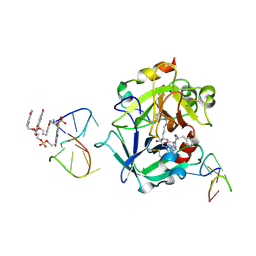 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form delta | | Descriptor: | 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
7ZKN
 
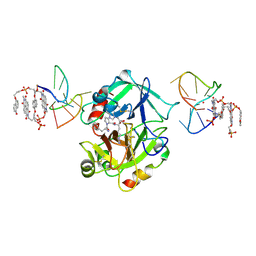 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form gamma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
8A0L
 
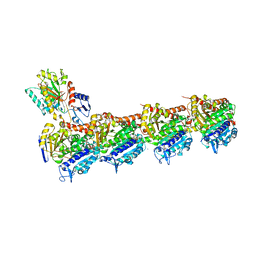 | | Tubulin-CW1-complex | | Descriptor: | (3~{S},4~{R},8~{S},10~{S},12~{S},14~{S})-14-[(~{Z},4~{R})-4-(hydroxymethyl)hex-2-en-2-yl]-4,12-dimethoxy-9,9-dimethyl-3,8,10-tris(oxidanyl)-1-oxacyclotetradecan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Diaz, J.F, Steinmetz, M.O, Oliva, M.A. | | Deposit date: | 2022-05-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9981 Å) | | Cite: | Chemical modulation of microtubule structure through the laulimalide/peloruside site.
Structure, 31, 2023
|
|
7ZKM
 
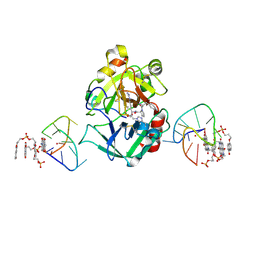 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
7ZKL
 
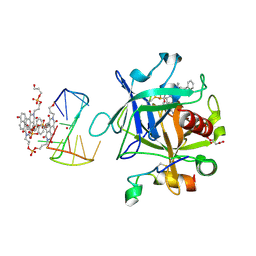 | | X-ray structure of the complex between human alpha thrombin and a pseudo-cyclic thrombin binding aptamer (TBA-NNp/DDp) - Crystal form alpha | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 3-[13-methyl-5,7,12,14-tetrakis(oxidanylidene)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),2,4(16),8,10-pentaen-6-yl]propyl 3-[5,7,12,14-tetrakis(oxidanylidene)-13-(3-oxidanylpropyl)-6,13-diazatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1,3,8(16),9,11(15)-pentaen-6-yl]propyl hydrogen phosphate, 3-[5-[3-bis(oxidanyl)phosphanyloxypropoxy]naphthalen-1-yl]oxypropyl 3-(5-oxidanylnaphthalen-1-yl)oxypropyl hydrogen phosphate, ... | | Authors: | Troisi, R, Sica, F. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | A terminal functionalization strategy reveals unusual binding abilities of anti-thrombin anticoagulant aptamers.
Mol Ther Nucleic Acids, 30, 2022
|
|
7ZZG
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},24~{R},25~{S},26~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-25,26-bis(oxidanyl)-27-oxa-2,4,6,9,14,17,20,22-octazatetracyclo[22.2.1.0^{2,10}.0^{3,8}]heptacosa-3(8),4,6,9-tetraen-11-yne-16,21-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
7ZLI
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZZH
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},22~{R},23~{S},24~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-23,24-bis(oxidanyl)-25-oxa-2,4,6,9,14,17,20-heptazatetracyclo[20.2.1.0^{2,10}.0^{3,8}]pentacosa-3(8),4,6,9-tetraen-11-yne-16,19-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
7ZZJ
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},23~{R},24~{S},25~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-24,25-bis(oxidanyl)-26-oxa-2,4,6,9,14,17,21-heptazatetracyclo[21.2.1.0^{2,10}.0^{3,8}]hexacosa-3(8),4,6,9-tetraen-11-yne-16,20-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
