3WI3
 
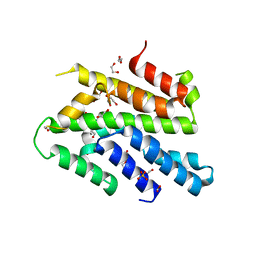 | | Crystal Structure of the Sld3/Treslin domain from yeast Sld3 | | Descriptor: | 1,2-ETHANEDIOL, DNA replication regulator SLD3, SULFATE ION | | Authors: | Itou, H, Araki, H, Shirakihara, Y. | | Deposit date: | 2013-09-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the homology domain of the eukaryotic DNA replication proteins sld3/treslin.
Structure, 22, 2014
|
|
4O8Y
 
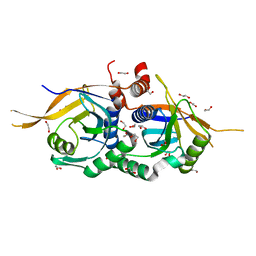 | | Zinc-free Rpn11 in complex with Rpn8 | | Descriptor: | 1,2-ETHANEDIOL, 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN8 | | Authors: | Worden, E.J, Padovani, C, Martin, A. | | Deposit date: | 2013-12-30 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4GFT
 
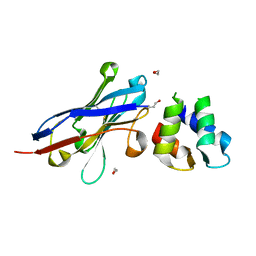 | | Malaria invasion machinery protein-Nanobody complex | | Descriptor: | 1,2-ETHANEDIOL, Myosin A tail domain interacting protein, Nanobody | | Authors: | Khamrui, S, Turley, S, Pardon, E, Steyaert, J, Verlinde, C, Fan, E, Bergman, L.W, Hol, W.G.J. | | Deposit date: | 2012-08-03 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a nanobody.
Mol.Biochem.Parasitol., 190, 2013
|
|
3O12
 
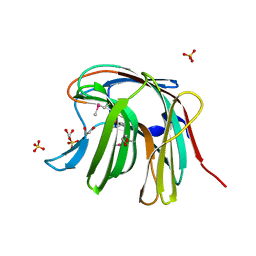 | | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein YJL217W | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae.
TO BE PUBLISHED
|
|
4OO0
 
 | |
1YSL
 
 | | Crystal structure of HMG-CoA synthase from Enterococcus faecalis with AcetoAcetyl-CoA ligand. | | Descriptor: | ACETOACETIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Steussy, C.N, Vartia, A.A, Burgner II, J.W, Sutherlin, A, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2005-02-08 | | Release date: | 2005-11-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Crystal Structures of HMG-CoA Synthase from Enterococcus faecalis and a Complex with Its Second Substrate/Inhibitor Acetoacetyl-CoA.
Biochemistry, 44, 2005
|
|
4GG7
 
 | | Crystal structure of cMET in complex with novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-(3-nitrobenzyl)-6-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-2-(trifluoromethyl)pyrido[2,3-d]pyrimidin-4-amine | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6MDX
 
 | | Mechanism of protease dependent DPC repair | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DNA (5'-D(P*CP*C)-3'), ... | | Authors: | Li, F, Raczynska, J, Chen, Z, Yu, H. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-10 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insight into DNA-Dependent Activation of Human Metalloprotease Spartan.
Cell Rep, 26, 2019
|
|
3O8U
 
 | |
6M77
 
 | |
4GB7
 
 | | Putative 6-aminohexanoate-dimer hydrolase from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, 6-aminohexanoate-dimer hydrolase, NITRATE ION | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-26 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Putative 6-aminohexanoate-dimer hydrolase from Bacillus anthracis.
To be Published
|
|
1XXZ
 
 | | Solution structure of sst1-selective somatostatin (SRIF) analog | | Descriptor: | SST1-selective somatosatin (analog 5) | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
1XSV
 
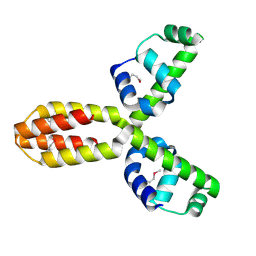 | | X-ray crystal structure of conserved hypothetical UPF0122 protein SAV1236 from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | Hypothetical UPF0122 protein SAV1236 | | Authors: | Walker, J.R, Xu, X, Virag, C, McDonald, M.-L, Houston, S, Buzadzija, K, Vedadi, M, Dharamsi, A, Fiebig, K.M, Savchenko, A. | | Deposit date: | 2004-10-20 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Crystal Structure of Conserved Hypothetical UPF0122 Protein SAV1236 From Staphylococcus aureus
To be Published
|
|
2N76
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | Descriptor: | De novo designed protein LFR1 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414
To be Published
|
|
9OG5
 
 | | SARS-COV-2-6P-MUT7 S PROTEIN-DY-III-281 complex 1 RBD up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Chandravanshi, M, Niu, L, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2025-04-30 | | Release date: | 2025-10-01 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of VE607 to generate analogs with improved neutralization activities against SARS-CoV-2 variants.
J.Virol., 2025
|
|
4PMY
 
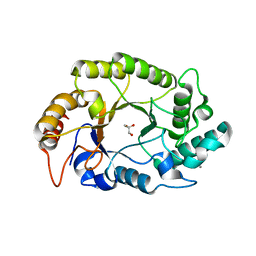 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylose | | Descriptor: | CALCIUM ION, GLYCEROL, Xylanase, ... | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
1LSU
 
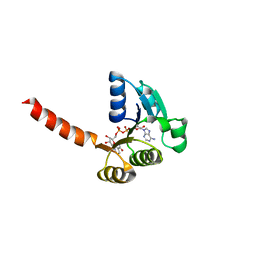 | | KTN Bsu222 Crystal Structure in Complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Conserved hypothetical protein yuaA | | Authors: | Roosild, T.P, Miller, S, Booth, I.R, Choe, S. | | Deposit date: | 2002-05-18 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch.
Cell(Cambridge,Mass.), 109, 2002
|
|
8QHR
 
 | | Crystal structure of the human DNPH1 glycosyl-enzyme intermediate | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, ... | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
6S41
 
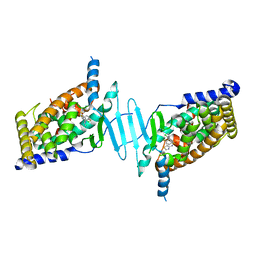 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH XPC-7455 | | Descriptor: | 4-[[(1~{S})-1-[2,5-bis(fluoranyl)phenyl]ethyl]amino]-5-chloranyl-2-fluoranyl-~{N}-(1,3-thiazol-4-yl)benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Focken, T, Maskos, K, Griessner, A, Krapp, S. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of CNS-Penetrant Aryl Sulfonamides as Isoform-Selective NaV1.6 Inhibitors with Efficacy in Mouse Models of Epilepsy.
J.Med.Chem., 62, 2019
|
|
9BFP
 
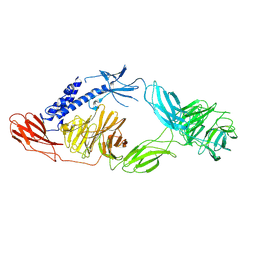 | | Cryo-EM structure of Sevenless extracellular domain (monomer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein sevenless | | Authors: | Cerutti, G, Shapiro, L. | | Deposit date: | 2024-04-18 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structures and pH-dependent dimerization of the sevenless receptor tyrosine kinase.
Mol.Cell, 84, 2024
|
|
6PNP
 
 | | Crystal structure of the splice insert-free neurexin-1 LNS2 domain in complex with neurexophilin-1 | | Descriptor: | Neurexin-1, Neurexophilin-1 | | Authors: | Wilson, S.C, White, K.I, Zhou, Q, Brunger, A.T. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of neurexophilin-neurexin complexes reveal a regulatory mechanism of alternative splicing.
Embo J., 38, 2019
|
|
3KC6
 
 | |
7PEM
 
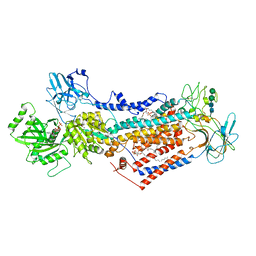 | | Cryo-EM structure of phophorylated Drs2p-Cdc50p in a PS and ATP-bound E2P state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Timcenko, M, Wang, Y, Lyons, J.A, Nissen, P, Lindorff-Larsen, K. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Substrate Transport and Specificity in a Phospholipid Flippase
To Be Published
|
|
1HU5
 
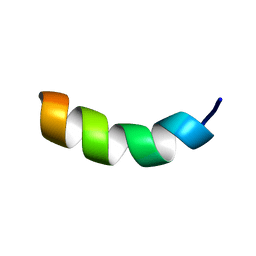 | | SOLUTION STRUCTURE OF OVISPIRIN-1 | | Descriptor: | OVISPIRIN-1 | | Authors: | Sawai, M.V, Waring, A.J, Kearney, W.R, McCray Jr, P.B, Forsyth, W.R, Lehrer, R.I, Tack, B.F. | | Deposit date: | 2001-01-04 | | Release date: | 2002-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Impact of single-residue mutations on the structure and function of ovispirin/novispirin antimicrobial peptides.
Protein Eng., 15, 2002
|
|
9QEG
 
 | | Cryo-EM structure of the 70S ribosome of a MLSb sensitive S. aureus strain "KES34" in complex with solithromycin | | Descriptor: | (3aS,4R,7S,9R,10R,11R,13R,15R,15aR)-1-{4-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]butyl}-4-ethyl-7-fluoro-11-methoxy-3a ,7,9,11,13,15-hexamethyl-2,6,8,14-tetraoxotetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-10-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Rivalta, A, Yonath, A. | | Deposit date: | 2025-03-10 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural studies on ribosomes of differentially macrolide-resistant Staphylococcus aureus strains.
Life Sci Alliance, 8, 2025
|
|
