6P1C
 
 | |
8C8J
 
 | | Long Interspersed Nuclear Element 1 (LINE-1) reverse transcriptase ternary complex with hybrid duplex and dTTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, CHLORIDE ION, ... | | Authors: | Nichols, C.E, Walpole, T.B, Baldwin, E. | | Deposit date: | 2023-01-20 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
7V2W
 
 | | protomer structure from the dimer of yeast THO complex | | Descriptor: | Protein TEX1, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
1KDH
 
 | | Binary Complex of Murine Terminal Deoxynucleotidyl Transferase with a Primer Single Stranded DNA | | Descriptor: | 5'-D(P*(BRU)P*(BRU)P*(BRU)P*(BRU))-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Jourdan, N, Sukumar, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-11-13 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
EMBO J., 21, 2002
|
|
5O1T
 
 | |
1GRJ
 
 | |
6KR6
 
 | | Crystal structure of Drosophila Piwi | | Descriptor: | MERCURY (II) ION, Protein piwi, ZINC ION, ... | | Authors: | Yamaguchi, S, Oe, A, Yamashita, K, Hirano, S, Mastumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Drosophila Piwi.
Nat Commun, 11, 2020
|
|
5U0P
 
 | | Cryo-EM structure of the transcriptional Mediator | | Descriptor: | Mediator complex subunit 10, Mediator complex subunit 11, Mediator complex subunit 14, ... | | Authors: | Tsai, K.-L, Yu, X, Gopalan, S, Chao, T.-C, Zhang, Y, Florens, L, Washburn, M.P, Murakami, K, Conaway, R.C, Conaway, J.W, Asturias, F. | | Deposit date: | 2016-11-26 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mediator structure and rearrangements required for holoenzyme formation.
Nature, 544, 2017
|
|
2HHW
 
 | | ddTTP:O6-methyl-guanine pair in the polymerase active site, in the closed conformation | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6WS5
 
 | | Rational drug design of phenazopyridine derivatives as novel inhibitors of Rev1-CT | | Descriptor: | 3-[(Z)-(2,3-difluorophenyl)diazenyl]pyridine-2,6-diamine, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | McPherson, K.S, Korzhnev, D.M. | | Deposit date: | 2020-04-30 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structure-Based Drug Design of Phenazopyridine Derivatives as Inhibitors of Rev1 Interactions in Translesion Synthesis.
Chemmedchem, 16, 2021
|
|
8FY2
 
 | | E3:PROTAC:target ternary complex structure (VCB/WH244/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Ruben, E, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY0
 
 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-xL) | | Descriptor: | Bcl-2-like protein 1, CACODYLIC ACID, Elongin-B, ... | | Authors: | Olsen, S.K, Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY1
 
 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
5W2E
 
 | | HCV NS5B RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor MK-8876 | | Descriptor: | 2-(4-fluorophenyl)-5-(11-fluoro-6H-pyrido[2',3':5,6][1,3]oxazino[3,4-a]indol-2-yl)-N-methyl-6-[methyl(methylsulfonyl)amino]-1-benzofuran-3-carboxamide, Genome polyprotein | | Authors: | Lesburg, C.A, Ummat, A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a New Structural Class of Broadly Acting HCV Non-Nucleoside Inhibitors Leading to the Discovery of MK-8876.
ChemMedChem, 12, 2017
|
|
6T9I
 
 | | cryo-EM structure of transcription coactivator SAGA | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6WS0
 
 | |
5O1X
 
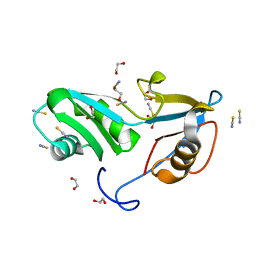 | | Structure of Nrd1 RNA binding domain | | Descriptor: | 1,2-ETHANEDIOL, Protein NRD1, THIOCYANATE ION | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
7U5C
 
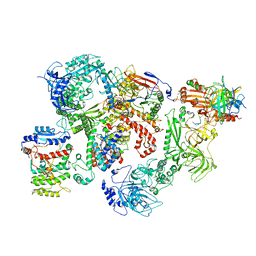 | | Cryo-EM structure of human CST bound to DNA polymerase alpha-primase in a recruitment state | | Descriptor: | CST complex subunit CTC1, CST complex subunit STN1, CST complex subunit TEN1, ... | | Authors: | Cai, S.W, Zinder, J.C, Svetlov, V, Bush, M.W, Nudler, E, Walz, T, de Lange, T. | | Deposit date: | 2022-03-02 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structure of the human CST-Pol alpha /primase complex in a recruitment state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5O1W
 
 | |
6T9K
 
 | | SAGA Core module | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6SO2
 
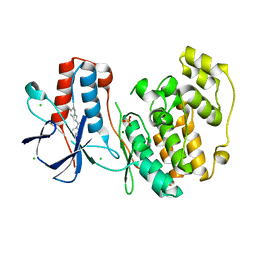 | | Fragment N13460a in complex with MAP kinase p38-alpha | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mitogen-activated protein kinase 14, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-08-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6SPL
 
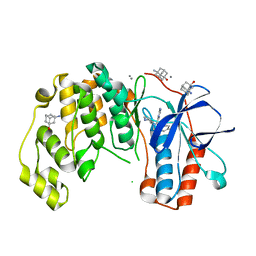 | | Fragment KCL615 in complex with MAP kinase p38-alpha | | Descriptor: | (5~{S},7~{R})-3-azanyladamantan-1-ol, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, CALCIUM ION, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6SO1
 
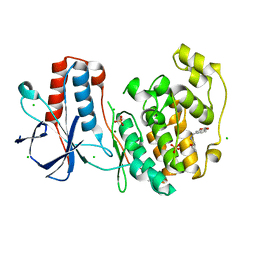 | | Fragment N13569a in complex with MAP kinase p38-alpha | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-[[(2~{R})-oxolan-2-yl]methyl]methanamine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-08-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6SOI
 
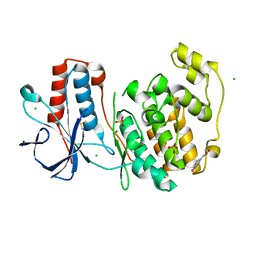 | | Fragment N13788a in complex with MAP kinase p38-alpha | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6SOT
 
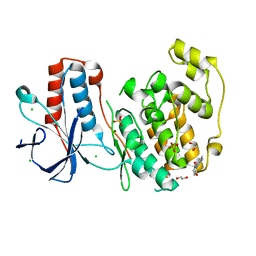 | | Fragment N11290a in complex with MAP kinase p38-alpha | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methylphenyl)pyrrolidine-2,5-dione, CHLORIDE ION, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
