3H4C
 
 | | Structure of the C-terminal Domain of Transcription Factor IIB from Trypanosoma brucei | | Descriptor: | 1,2-ETHANEDIOL, Transcription factor TFIIB-like | | Authors: | Syed Ibrahim, B, Kanneganti, N, Rieckhof, G.E, Das, A, Laurents, D.V, Palenchar, J.B, Bellofatto, V, Wah, D.A. | | Deposit date: | 2009-04-18 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of transcription factor IIB from Trypanosoma brucei.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8CG9
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with a cyclic N-hydroxyurea inhibitor | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, 1-[[(3~{R})-2,3-dihydro-1,4-benzodioxin-3-yl]methyl]-3-oxidanyl-thieno[3,2-d]pyrimidine-2,4-dione, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
4EPZ
 
 | |
4NEO
 
 | | Structure of BlmI, a type-II acyl-carrier-protein from Streptomyces verticillus involved in bleomycin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Peptide synthetase NRPS type II-PCP | | Authors: | Cuff, M.E, Bigelow, L, Bearden, J, Babnigg, G, Bruno, C.J.P, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of BlmI as a model for nonribosomal peptide synthetase peptidyl carrier proteins.
Proteins, 82, 2014
|
|
5BN3
 
 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ADP from Nanoarcheaum equitans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | Deposit date: | 2015-05-25 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
7WAR
 
 | |
6FO5
 
 | | FIRST DOMAIN OF HUMAN BROMODOMAIN BRD4 IN COMPLEX WITH INHIBITOR #17 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[[4-[[7-ethyl-2,6-bis(oxidanylidene)purin-3-yl]methyl]phenyl]methyl]-2-oxidanylidene-1,3,4,5-tetrahydro-1-benzazepine-7-sulfonamide | | Authors: | Raux, B, Betzi, S. | | Deposit date: | 2018-02-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Integrated Strategy for Lead Optimization Based on Fragment Growing: The Diversity-Oriented-Target-Focused-Synthesis Approach.
J. Med. Chem., 61, 2018
|
|
8QWL
 
 | | Structure of p53 cancer mutant Y163C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, MALONATE ION, ... | | Authors: | Balourdas, D.I, Markl, A.M, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
6DRR
 
 | | Crystal structure of Cj0485 dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, Short-chain dehydrogenase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2018-06-12 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The gastrointestinal pathogen Campylobacter jejuni metabolizes sugars with potential help from commensal Bacteroides vulgatus.
Commun Biol, 3, 2020
|
|
5J1U
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-(furan-3-ylmethyl)pyrrolidine-1-carboxamide at 1.80A resolution | | Descriptor: | EthR, N-[(furan-3-yl)methyl]pyrrolidine-1-carboxamide | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
8VOJ
 
 | | The Cryo-EM structure of LSD1-CoREST-HDAC1 in complex with KBTBD4 enhanced by UM171 and IP6 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Xie, X, Mao, H, Liau, B, Zheng, N. | | Deposit date: | 2024-01-15 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | UM171 glues asymmetric CRL3-HDAC1/2 assembly to degrade CoREST corepressors.
Nature, 639, 2025
|
|
7WIK
 
 | |
6JNT
 
 | | Catalase structure determined by eEFD (dataset 1) | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yonekura, K, Maki-Yonekura, S. | | Deposit date: | 2019-03-18 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | A new cryo-EM system for electron 3D crystallography by eEFD.
J.Struct.Biol., 206, 2019
|
|
7TH2
 
 | | Single-domain VHH intrabodies neutralize ricin toxin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Single-domain antibodies neutralize ricin toxin intracellularly by blocking access to ribosomal P-stalk proteins.
J.Biol.Chem., 298, 2022
|
|
6G54
 
 | | Crystal structure of ERK2 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 1, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
7WCP
 
 | |
6JQ7
 
 | |
6SQZ
 
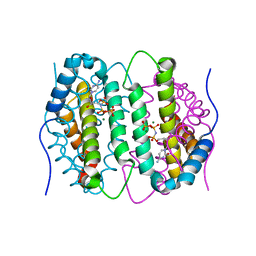 | | Mouse dCTPase in complex with dCMPNPP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
4YU9
 
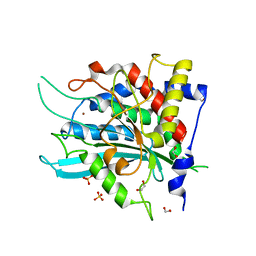 | | Crystal Structure of double mutant Y115E Y117E human Glutaminyl Cyclase | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Di Pisa, F, Pozzi, C, Benvenuti, M, Mangani, S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The soluble Y115E-Y117E variant of human glutaminyl cyclase is a valid target for X-ray and NMR screening of inhibitors against Alzheimer disease.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5J7C
 
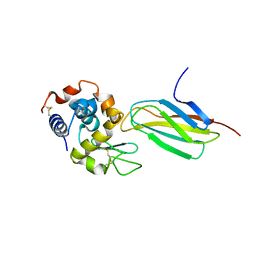 | |
6D8V
 
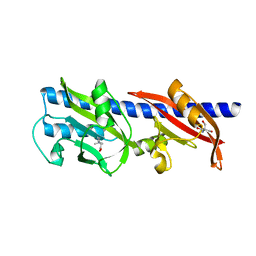 | | Methyl-accepting Chemotaxis protein X | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Probable chemoreceptor (Methyl-accepting chemotaxis) transmembrane protein | | Authors: | Shrestha, M, Schubot, F.D. | | Deposit date: | 2018-04-27 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the sensory domain of McpX fromSinorhizobium meliloti, the first known bacterial chemotactic sensor for quaternary ammonium compounds.
Biochem. J., 475, 2018
|
|
7ZQT
 
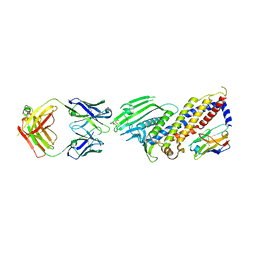 | |
5J7Q
 
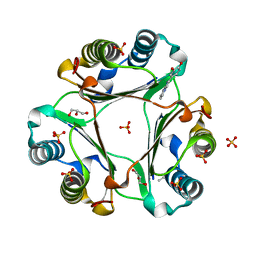 | | Macrophage Migration Inhibitory Factor bound to Inhibitor K664 Derivative | | Descriptor: | 4-(imidazo[1,2-a]pyridin-2-yl)benzene-1,2-diol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Irregularities in enzyme assays: The case of macrophage migration inhibitory factor.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8HNN
 
 | | Structure of CXCR3 complexed with antagonist SCH546738 | | Descriptor: | 3-azanyl-6-chloranyl-5-[(3S)-4-[1-[(4-chlorophenyl)methyl]piperidin-4-yl]-3-ethyl-piperazin-1-yl]pyrazine-2-carboxamide, CHOLESTEROL, Nb6, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9F2V
 
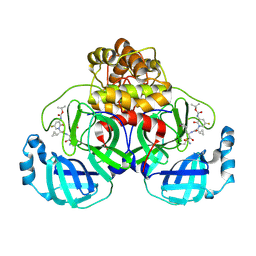 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR02 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{R},3~{S})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-5-fluoranyl-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-24 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
