5HLS
 
 | | Crystal structure of the first bromodomain of human BRD4 bound to CPI-0610 | | Descriptor: | Bromodomain-containing protein 4, CPI-0610 | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-01-15 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Identification of a Benzoisoxazoloazepine Inhibitor (CPI-0610) of the Bromodomain and Extra-Terminal (BET) Family as a Candidate for Human Clinical Trials.
J.Med.Chem., 59, 2016
|
|
1NZ7
 
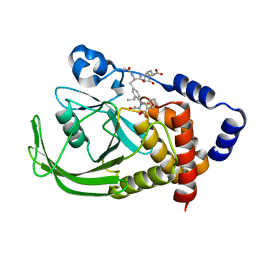 | | POTENT, SELECTIVE INHIBITORS OF PROTEIN TYROSINE PHOSPHATASE 1B USING A SECOND PHOSPHOTYROSINE BINDING SITE, complexed with compound 19. | | Descriptor: | 2-[(4-{2-ACETYLAMINO-2-[4-(1-CARBOXY-3-METHYLSULFANYL-PROPYLCARBAMOYL)-BUTYLCARBAMOYL]-ETHYL}-2-ETHYL-PHENYL)-OXALYL-AM INO]-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Xin, Z, Oost, T.K, Abad-Zapatero, C, Hajduk, P.J, Pei, Z, Szczepankiewicz, B.G, Hutchins, C.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Trevillyan, J.M, Jirousek, M.R, Liu, G. | | Deposit date: | 2003-02-16 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent, Selective Inhibitors of Protein Tyrosine Phosphatase 1B
BIOORG.MED.CHEM.LETT., 13, 2003
|
|
4AF3
 
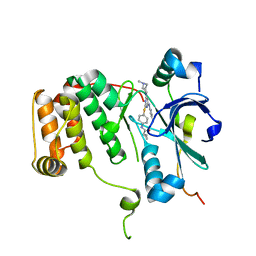 | | Human Aurora B Kinase in complex with INCENP and VX-680 | | Descriptor: | AURORA KINASE B, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, INNER CENTROMERE PROTEIN | | Authors: | Elkins, J.M, Vollmar, M, Wang, J, Picaud, S, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Knapp, S. | | Deposit date: | 2012-01-16 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Human Aurora B in Complex with Incenp and Vx-680.
J.Med.Chem., 55, 2012
|
|
1QD8
 
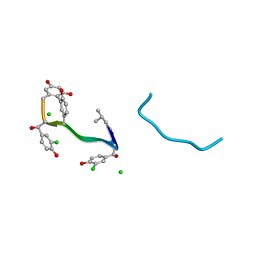 | | COMPLEX OF VANCOMYCIN WITH N-ACETYL GLYCINE | | Descriptor: | ACETYLAMINO-ACETIC ACID, CHLORIDE ION, VANCOMYCIN, ... | | Authors: | Loll, P.J, Kaplan, J, Selinsky, B, Axelsen, P.H. | | Deposit date: | 1999-07-15 | | Release date: | 1999-08-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Vancomycin Binding to Low-Affinity Ligands: Delineating a Minimum Set of Interactions Necessary for High-Affinity Binding.
J.Med.Chem., 42, 1999
|
|
4BCG
 
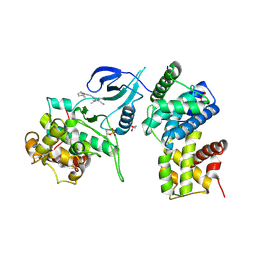 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[[3-(1,4-diazepan-1-yl)phenyl]amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-04-17 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.085 Å) | | Cite: | Substituted 4-(Thiazol-5-Yl)-2-(Phenylamino)Pyrimidines are Highly Active Cdk9 Inhibitors: Synthesis, X-Ray Crystal Structure, Sar and Anti-Cancer Activities.
J.Med.Chem., 56, 2013
|
|
4BCP
 
 | | Structure of CDK2 in complex with cyclin A and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[[3-(1,4-diazepan-1-yl)phenyl]amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-04-17 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Substituted 4-(Thiazol-5-Yl)-2-(Phenylamino)Pyrimidines are Highly Active Cdk9 Inhibitors: Synthesis, X-Ray Crystal Structure, Sar and Anti-Cancer Activities.
J.Med.Chem., 56, 2013
|
|
2ANO
 
 | | Crystal structure of E.coli dihydrofolate reductase in complex with NADPH and the inhibitor MS-SH08-17 | | Descriptor: | 1-{[N-(1-IMINO-GUANIDINO-METHYL)]SULFANYLMETHYL}-3-TRIFLUOROMETHYL-BENZENE, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
5HMZ
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 23 | | Descriptor: | 5-(5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl)-4-methoxy-2-methyl-N-(methylsulfonyl)benzamide, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design
J.Med.Chem., 59, 2016
|
|
5HMX
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 10 | | Descriptor: | 2,2'-(5-(thiophen-2-yl)-1,3-phenylene)diacetic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design
J.Med.Chem., 59, 2016
|
|
5HMY
 
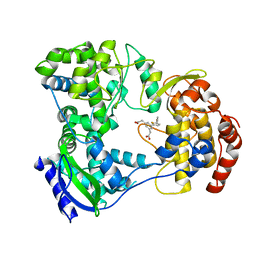 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 15 | | Descriptor: | 2,2'-(5-(5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl)-1,3-phenylene)diacetic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design.
J.Med.Chem., 59, 2016
|
|
6DUM
 
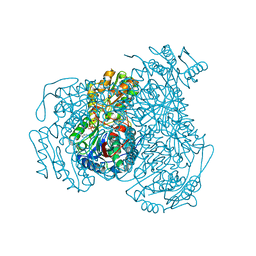 | | ALDH1A1 N121S in complex with 6-{[(3-fluorophenyl)methyl]sulfanyl}-2-(oxetan-3-yl)-5-phenyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one (compound 13g) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 6-{[(3-fluorophenyl)methyl]sulfanyl}-2-(oxetan-3-yl)-5-phenyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, ... | | Authors: | Buchman, C.D, Hurley, T.D. | | Deposit date: | 2018-06-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Optimization of a Novel Class of Aldehyde Dehydrogenase 1A (ALDH1A) Subfamily-Selective Inhibitors as Potential Adjuncts to Ovarian Cancer Chemotherapy.
J.Med.Chem., 61, 2018
|
|
5HMW
 
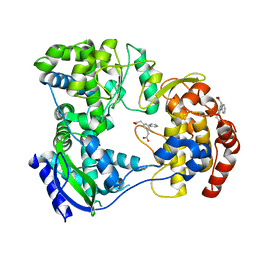 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 5 | | Descriptor: | 2,2'-biphenyl-3,5-diyldiacetic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design
J.Med.Chem., 59, 2016
|
|
6R5H
 
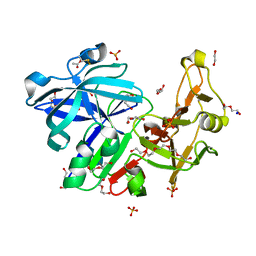 | | Major aspartyl peptidase 1 from C. neoformans | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Krystufek, R, Sacha, P, Brynda, J, Konvalinka, J. | | Deposit date: | 2019-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2021-06-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery.
J.Med.Chem., 64, 2021
|
|
6R6A
 
 | | Major aspartyl peptidase 1 from C. neoformans | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Endopeptidase, ... | | Authors: | Krystufek, R, Sacha, P, Brynda, J, Konvalinka, J. | | Deposit date: | 2019-03-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery.
J.Med.Chem., 64, 2021
|
|
5I40
 
 | | BRD9 in complex with Cpd1 (6-methyl-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one) | | Descriptor: | 1,2-ETHANEDIOL, 6-methyl-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Bromodomain-containing protein 9, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.0402 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
5I7Y
 
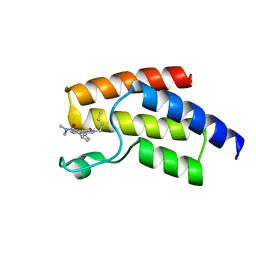 | | BRD9 in complex with Cpd4 ((E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide) | | Descriptor: | 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Bromodomain-containing protein 9 | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4514 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
4N96
 
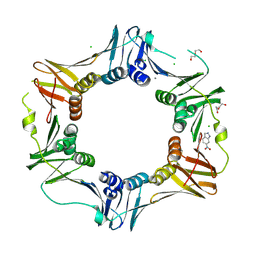 | | E. coli sliding clamp in complex with 6-nitroindazole | | Descriptor: | 6-NITROINDAZOLE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N95
 
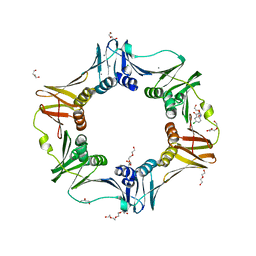 | | E. coli sliding clamp in complex with 5-chloroindoline-2,3-dione | | Descriptor: | 5-chloro-1H-indole-2,3-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
5I7X
 
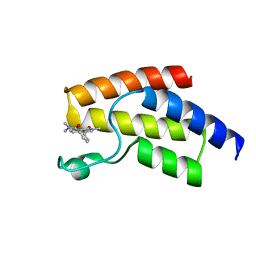 | | BRD9 in complex with Cpd2 (N,N-dimethyl-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)benzamide) | | Descriptor: | Bromodomain-containing protein 9, N,N-dimethyl-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)benzamide | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1752 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
4N98
 
 | |
6PYS
 
 | | Human PI3Kalpha in complex with Compound 2-10 ((3S)-3-benzyl-3-methyl-5-[5-(2-methylpyrimidin-5-yl)pyrazolo[1,5-a]pyrimidin-3-yl]-1,3-dihydro-2H-indol-2-one) | | Descriptor: | (3S)-3-benzyl-3-methyl-5-[5-(2-methylpyrimidin-5-yl)pyrazolo[1,5-a]pyrimidin-3-yl]-1,3-dihydro-2H-indol-2-one, GLYCEROL, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Lesburg, C.A, Augustin, M.A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Design of selective PI3K delta inhibitors using an iterative scaffold-hopping workflow.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5JSF
 
 | | Crystal structure of 17beta-hydroxysteroid dehydrogenase 14 S205 variant in complex with NAD. | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-05-08 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | New Insights into Human 17 beta-Hydroxysteroid Dehydrogenase Type 14: First Crystal Structures in Complex with a Steroidal Ligand and with a Potent Nonsteroidal Inhibitor.
J.Med.Chem., 59, 2016
|
|
4N9A
 
 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid | | Descriptor: | (1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N97
 
 | | E. coli sliding clamp in complex with 5-nitroindole | | Descriptor: | 5-nitro-1H-indole, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
7U30
 
 | | PRMT5:MEP50 Complexed with Cyclonucleoside Compound 1 | | Descriptor: | (9R,10R,11S,12R,13R,14R)-4-amino-9-(3,4-difluorophenyl)-6,7,8,9,10,11,12,13-octahydro-10,13-epoxy[1,3]diazecino[1,2-e]purine-11,12-diol, 1,2-ETHANEDIOL, Methylosome protein 50, ... | | Authors: | Palte, R.L. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design and synthesis of unprecedented 9- and 10-membered cyclonucleosides with PRMT5 inhibitory activity.
Bioorg.Med.Chem., 66, 2022
|
|
