1ZAU
 
 | |
1TA8
 
 | |
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
1IK9
 
 | | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | | Descriptor: | DNA LIGASE IV, DNA REPAIR PROTEIN XRCC4 | | Authors: | Sibanda, B.L, Critchlow, S.E, Begun, J, Pei, X.Y, Jackson, S.P, Blundell, T.L, Pellegrini, L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an Xrcc4-DNA ligase IV complex.
Nat.Struct.Biol., 8, 2001
|
|
4GLW
 
 | | DNA ligase A in complex with inhibitor | | Descriptor: | 7-methoxy-6-methylpteridine-2,4-diamine, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Prade, L, Lange, R, Tidten-Luksch, N, Chambovey, A. | | Deposit date: | 2012-08-15 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design, synthesis and biological evaluation of novel DNA ligase inhibitors with in vitro and in vivo anti-staphylococcal activity.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6KKV
 
 | |
6WH1
 
 | | Structure of the complex of human DNA ligase III-alpha and XRCC1 BRCT domains | | Descriptor: | DNA ligase 3 alpha, X-ray repair cross complementing protein 1 variant | | Authors: | Pourfarjam, Y, Ellenberger, T, Tainer, J.A, Tomkinson, A.E, Kim, I.K. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex.
Nucleic Acids Res., 49, 2021
|
|
4D05
 
 | | Structure and activity of a minimal-type ATP-dependent DNA ligase from a psychrotolerant bacterium | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT DNA LIGASE, MAGNESIUM ION, ... | | Authors: | Williamson, A, Rothweiler, U, Leiros, H.-K.S. | | Deposit date: | 2014-04-24 | | Release date: | 2014-11-12 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enzyme-Adenylate Structure of a Bacterial ATP-Dependent DNA Ligase with a Minimized DNA-Binding Surface
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3PN1
 
 | | Novel Bacterial NAD+-dependent DNA Ligase Inhibitors with Broad Spectrum Potency and Antibacterial Efficacy In Vivo | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-(butylsulfanyl)adenosine, DNA ligase | | Authors: | Mills, S, Eakin, A, Buurman, E, Newman, J, Gao, N, Huynh, H, Johnson, K, Lahiri, S, Shapiro, A, Walkup, G, Wei, Y, Stokes, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Bacterial NAD+-Dependent DNA Ligase Inhibitors with Broad-Spectrum Activity and Antibacterial Efficacy In Vivo.
Antimicrob.Agents Chemother., 55, 2011
|
|
3VNN
 
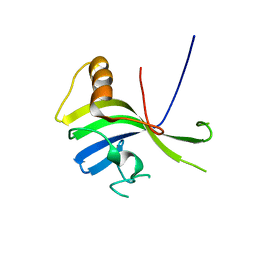 | | Crystal Structure of a sub-domain of the nucleotidyltransferase (adenylation) domain of human DNA ligase IV | | Descriptor: | DNA ligase 4 | | Authors: | Ochi, T, Wu, Q, Chirgadze, D.Y, Grossmann, J.G, Bolanos-Garcia, V.M, Blundell, T.L. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural insights into the role of domain flexibility in human DNA ligase IV
Structure, 20, 2012
|
|
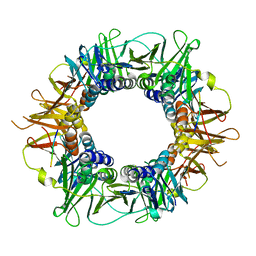
5FRQ
 
 | |
1IN1
 
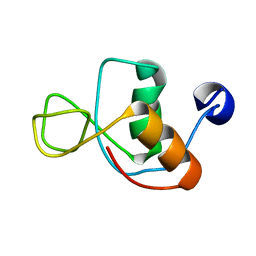 | |
1IMO
 
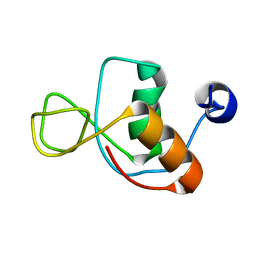 | |
2OWO
 
 | | Last Stop on the Road to Repair: Structure of E.coli DNA Ligase Bound to Nicked DNA-Adenylate | | Descriptor: | 26-MER, 5'-D(*AP*CP*AP*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*C)-3', 5'-D(*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*G)-3', ... | | Authors: | Shuman, S, Nandakumar, J, Nair, P.A. | | Deposit date: | 2007-02-16 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Last Stop on the Road to Repair: Structure of E. coli DNA Ligase Bound to Nicked DNA-Adenylate.
Mol.Cell, 26, 2007
|
|
6KRH
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis NAD + -dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6KSD
 
 | |
6KSC
 
 | |
6LW8
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | (4R)-4-(4-fluorophenyl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine, DNA ligase A, GLYCEROL, ... | | Authors: | Ramachandran, R, Afsar, M, Shukla, A. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure based identification of first-in-class fragment inhibitors that target the NMN pocket of M. tuberculosis NAD + -dependent DNA ligase A.
J.Struct.Biol., 213, 2021
|
|
1P8L
 
 | | New Crystal Structure of Chlorella Virus DNA Ligase-Adenylate | | Descriptor: | ADENOSINE MONOPHOSPHATE, PBCV-1 DNA ligase | | Authors: | Odell, M, Malinina, L, Teplova, M, Shuman, S. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Analysis of the DNA Joining Repertoire of Chlorella Virus DNA ligase and a New Crystal Structure of the Ligase-Adenylate Intermediate
Nucleic Acids Res., 31, 2003
|
|
3W1G
 
 | | Crystal Structure of Human DNA ligase IV-Artemis Complex (Native) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Artemis-derived peptide, DNA ligase 4, ... | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2012-11-15 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the catalytic region of DNA ligase IV in complex with an artemis fragment sheds light on double-strand break repair
Structure, 21, 2013
|
|
3W1B
 
 | |
3W5O
 
 | | Crystal Structure of Human DNA ligase IV | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA ligase 4, SULFATE ION | | Authors: | Gu, X, Ochi, T, Blundell, T.L. | | Deposit date: | 2013-02-02 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of the catalytic region of DNA ligase IV in complex with an artemis fragment sheds light on double-strand break repair
Structure, 21, 2013
|
|
1X9N
 
 | | Crystal Structure of Human DNA Ligase I bound to 5'-adenylated, nicked DNA | | Descriptor: | 5'-phosphorylated DNA, ADENOSINE MONOPHOSPHATE, DNA ligase I, ... | | Authors: | Pascal, J.M, O'Brien, P.J, Tomkinson, A.E, Ellenberger, T. | | Deposit date: | 2004-08-23 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA ligase I completely encircles and partially unwinds nicked DNA.
Nature, 432, 2004
|
|
5WFY
 
 | |
4UFZ
 
 | | Synthesis of Novel NAD Dependant DNA Ligase Inhibitors via Negishi Cross-Coupling: Development of SAR and Resistance Studies | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5,7-bis(azanyl)-2-tert-butyl-4-(1,3-thiazol-2-yl)pyrido[2,3-d]pyrimidine-6-carbonitrile, DNA LIGASE | | Authors: | Murphy-Benenato, K.E, Boriack-Sjodin, P.A, Martinez-Botella, G, Carcanague, D, Gingipali, L, Gowravaram, M, Harang, J, Hale, M, Ioannidis, G, Jahic, H, Johnstone, M, Kutschke, A, Laganas, V.A, Loch, J, Oguto, H, Patel, S.J. | | Deposit date: | 2015-03-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Negishi Cross-Coupling Enabled Synthesis of Novel Nad(+)-Dependent DNA Ligase Inhibitors and Sar Development.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
