1S9V
 
 | | Crystal structure of HLA-DQ2 complexed with deamidated gliadin peptide | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DQ(1) beta chain, ... | | Authors: | Kim, C.-Y, Quarsten, H, Bergseng, E, Khosla, C, Sollid, L.M. | | Deposit date: | 2004-02-05 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7B2C
 
 | | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum gassed with xenon | | Descriptor: | (2R)-2-[(2S)-2-[(2S)-2-oxidanylpropoxy]propoxy]propan-1-ol, 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
3MHS
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
7B1S
 
 | | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum at 0.994-A resolution | | Descriptor: | (2S)-2-{[(2S)-2-{[(2S)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-25 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.992 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
1SL5
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with LNFP III (Dextra L504). | | Descriptor: | CALCIUM ION, MAGNESIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1S2V
 
 | | Crystal structure of phosphoenolpyruvate mutase complexed with Mg(II) | | Descriptor: | MAGNESIUM ION, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
6JU5
 
 | | Aspergillus oryzae pro-tyrosinase C92A/F513Y mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1SL4
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, mDC-SIGN1B type I isoform | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SQO
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 8-(PYRIMIDIN-2-YLAMINO)NAPHTHALENE-2-CARBOXIMIDAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-19 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1SME
 
 | | PLASMEPSIN II, A HEMOGLOBIN-DEGRADING ENZYME FROM PLASMODIUM FALCIPARUM, IN COMPLEX WITH PEPSTATIN A | | Descriptor: | PLASMEPSIN II, Pepstatin | | Authors: | Silva, A.M, Lee, A.Y, Gulnik, S.V, Goldberg, D.E, Erickson, J.W. | | Deposit date: | 1996-06-11 | | Release date: | 1997-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3MML
 
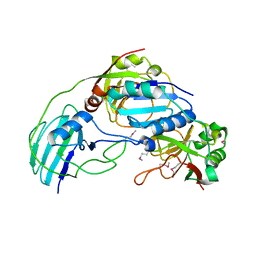 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
7AQS
 
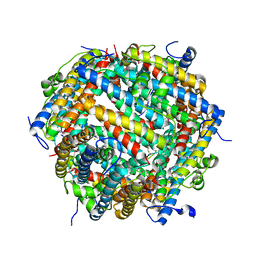 | | Crystal structure of E. coli DPS in space group P1 | | Descriptor: | DNA protection during starvation protein, FE (III) ION | | Authors: | Jakob, R.P, Pipercevic, J, Righetto, R, Goldie, K, Stahlberg, H, Maier, T, Hiller, S. | | Deposit date: | 2020-10-22 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a Dps contamination in Mitomycin-C-induced expression of Colicin Ia.
Biochim Biophys Acta Biomembr, 1863, 2021
|
|
3LZJ
 
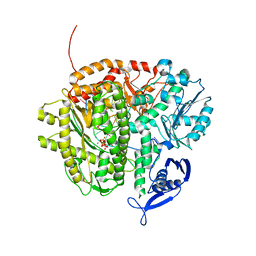 | | RB69 DNA Polymerase (Y567A) ternary complex with dCTP Opposite 7,8-Dihydro-8-oxoguanine | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-03-01 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine .
Biochemistry, 49, 2010
|
|
6JU9
 
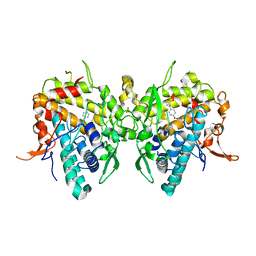 | | Aspergillus oryzae active-tyrosinase copper-bound C92A mutant complexed with L-tyrosine | | Descriptor: | 3,4-DIHYDROXYPHENYLALANINE, COPPER (II) ION, NITRATE ION, ... | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1SIF
 
 | | Crystal structure of a multiple hydrophobic core mutant of ubiquitin | | Descriptor: | ubiquitin | | Authors: | Benitez-Cardoza, C.G, Stott, K, Hirshberg, M, Went, H.M, Woolfson, D.N, Jackson, S.E. | | Deposit date: | 2004-02-29 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exploring sequence/folding space: folding studies on multiple hydrophobic core mutants of ubiquitin
Biochemistry, 43, 2004
|
|
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
1SLC
 
 | | X-RAY CRYSTALLOGRAPHY REVEALS CROSSLINKING OF MAMMALIAN LECTIN (GALECTIN-1) BY BIANTENNARY COMPLEX TYPE SACCHARIDES | | Descriptor: | BOVINE GALECTIN-1, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bourne, Y, Cambillau, C. | | Deposit date: | 1994-03-12 | | Release date: | 1995-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides.
Nat.Struct.Biol., 1, 1994
|
|
1S7Z
 
 | | Structure of Ocr from Bacteriophage T7 | | Descriptor: | CESIUM ION, Gene 0.3 protein | | Authors: | Walkinshaw, M.D, Taylor, P, Sturrock, S.S, Atanasiu, C, Berg, T, Henderson, R.M, Edwardson, J.M, Dryden, D.T. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of Ocr from Bacteriophage T7, a Protein that Mimics B-Form DNA
Mol.Cell, 9, 2002
|
|
3MEQ
 
 | | Crystal structure of alcohol dehydrogenase from Brucella melitensis | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase, ... | | Authors: | Arakaki, T.L, Staker, B.L, Gardberg, A, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Brucella melitensis
To be Published
|
|
3MHH
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
1SKS
 
 | | Binary 3' complex of T7 DNA polymerase with a DNA primer/template containing a cis-syn thymine dimer on the template | | Descriptor: | 5'-D(*CP*CP*CP*(TTD)P*AP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*AP*AP*CP*GP*AP*C*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*(2DT))-3', DNA polymerase, ... | | Authors: | Li, Y, Dutta, S, Doublie, S, Bdour, H.M, Taylor, J.S, Ellenberger, T. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nucleotide insertion opposite a cis-syn thymine dimer by a replicative DNA polymerase from bacteriophage T7.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1IXG
 
 | |
1SN5
 
 | | Crystal Structure of Sea Bream Transthyretin in complex with Triiodothyronine at 1.90A Resolution | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, SULFATE ION, transthyretin | | Authors: | Eneqvist, T, Lundberg, E, Karlsson, A, Huang, S, Cantos, C.R, Power, D.M, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of piscine transthyretin reveal different binding modes for triiodothyronine and thyroxine.
J.Biol.Chem., 279, 2004
|
|
1IXI
 
 | |
1SFO
 
 | | RNA POLYMERASE II STRAND SEPARATED ELONGATION COMPLEX | | Descriptor: | DNA STRAND, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Structural Basis of Transcription: Separation of RNA from DNA by RNA Polymerase II
Science, 303, 2004
|
|
