1F4C
 
 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE COVALENTLY MODIFIED AT C146 WITH N-[TOSYL-D-PROLINYL]AMINO-ETHANETHIOL | | Descriptor: | GLYCEROL, N-[TOSYL-D-PROLINYL]AMINO-ETHANETHIOL, SULFATE ION, ... | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
1F4L
 
 | | CRYSTAL STRUCTURE OF THE E.COLI METHIONYL-TRNA SYNTHETASE COMPLEXED WITH METHIONINE | | Descriptor: | METHIONINE, METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Serre, L, Verdon, G, Chonowski, T, Hervouet, N, Zelwer, C. | | Deposit date: | 2000-06-08 | | Release date: | 2001-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | How methionyl-tRNA synthetase creates its amino acid recognition pocket upon L-methionine binding.
J.Mol.Biol., 306, 2001
|
|
6UT6
 
 | | Cryo-EM structure of the Escherichia coli McrBC complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6S6A
 
 | | Crystal structure of RagA-Q66L/RagC-T90N GTPase heterodimer complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Anandapadamanaban, M, Masson, G.R, Perisic, O, Kaufman, J, Williams, R.L. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
6MEA
 
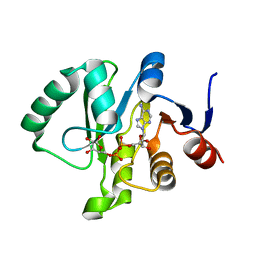 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate ribose (ADP-ribose) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6S6X
 
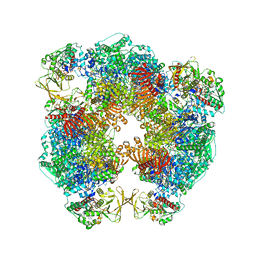 | | Structure of Azospirillum brasilense Glutamate Synthase in a6b6 oligomeric state. | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chaves-Sanjuan, A, Camilloni, C, Bolognesi, M. | | Deposit date: | 2019-07-03 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures of Azospirillum brasilense Glutamate Synthase in Its Oligomeric Assemblies.
J.Mol.Biol., 431, 2019
|
|
2MNZ
 
 | | NMR Structure of KDM5B PHD1 finger in complex with H3K4me0(1-10aa) | | Descriptor: | H3K4me0, Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
5CKM
 
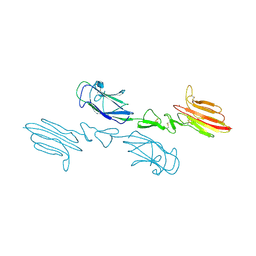 | | The CUB1-EGF-CUB2 domains of rat MBL-associated serine protease-2 (MASP-2) bound to Ca2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannan-binding lectin serine peptidase 2 | | Authors: | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | Deposit date: | 2015-07-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
8TWR
 
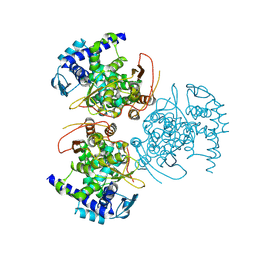 | | Influenza A virus (A/Aichi/2/1968(H3N2) nucleoprotein mutant - 2-7 deleted, P283S, R416A | | Descriptor: | Nucleoprotein, SODIUM ION | | Authors: | Yoon, J, Zhang, Y.M, Grant, R.A, Shoulders, M.D. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The immune-evasive proline-283 substitution in influenza nucleoprotein increases aggregation propensity without altering the native structure.
Sci Adv, 10, 2024
|
|
2LUU
 
 | | NMR solution structure of midkine-b, mdkb | | Descriptor: | Midkine-related growth factor Mdk2 | | Authors: | Yang, D, Lim, J, Meng, D. | | Deposit date: | 2012-06-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of full-length midkine reveals novel residues important for heparin binding and zebrafish embryogenesis.
Biochem.J., 451, 2013
|
|
1F35
 
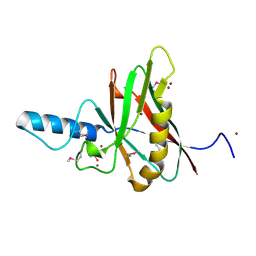 | |
3PPO
 
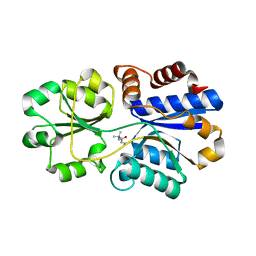 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | (2S)-3-carboxy-2-hydroxy-N,N,N-trimethylpropan-1-aminium, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
5GPE
 
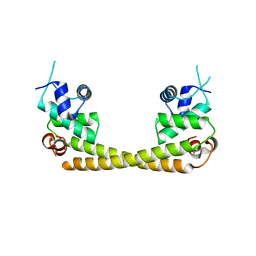 | | Crystal structure of the transcription regulator PbrR691 from Ralstonia metallidurans CH34 in complex with Lead(II) | | Descriptor: | LEAD (II) ION, Transcriptional regulator, MerR-family | | Authors: | Huang, S.Q, Chen, W.Z, Wang, D, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2016-08-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for the Selective Pb(II) Recognition of Metalloregulatory Protein PbrR691
Inorg Chem, 55, 2016
|
|
5CP6
 
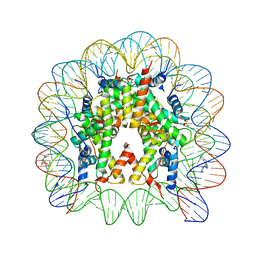 | | Nucleosome Core Particle with Adducts from the Anticancer Compound, [(eta6-5,8,9,10-tetrahydroanthracene)Ru(ethylenediamine)Cl][PF6] | | Descriptor: | (ethane6-5,8,9,10-tetrahydroanthracene)Ru(II)(ethylene-diamine)Cl, DNA (145-MER), Histone H2A, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2015-07-21 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Organometallic Compound which Exhibits a DNA Topology-Dependent One-Stranded Intercalation Mode.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7Q2U
 
 | | The crystal structure of the HINT1 Q62A mutant. | | Descriptor: | CACODYLATE ION, HEXAETHYLENE GLYCOL, Histidine triad nucleotide-binding protein 1, ... | | Authors: | Dolot, R.M, Strom, A.M, Wagner, C.R. | | Deposit date: | 2021-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Dynamic Long-Range Interactions Influence Substrate Binding and Catalysis by Human Histidine Triad Nucleotide-Binding Proteins (HINTs), Key Regulators of Multiple Cellular Processes and Activators of Antiviral ProTides.
Biochemistry, 61, 2022
|
|
1F4E
 
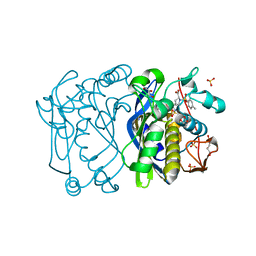 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE COMPLEXED WITH TOSYL-D-PROLINE | | Descriptor: | GLYCEROL, SULFATE ION, THYMIDYLATE SYNTHASE, ... | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
7VSU
 
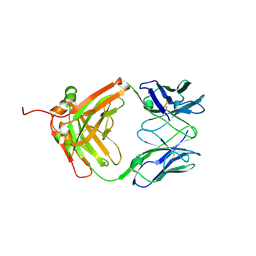 | |
5C71
 
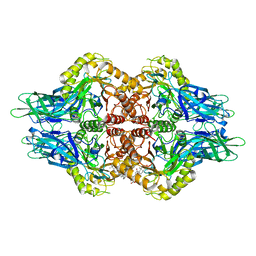 | | The structure of Aspergillus oryzae a-glucuronidase complexed with glycyrrhetinic acid monoglucuronide | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Glucuronidase, alpha-D-glucopyranuronic acid | | Authors: | Sun, H.L, Lv, B, Huang, S, Li, C, Jiang, T. | | Deposit date: | 2015-06-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure-guided engineering of the substrate specificity of a fungal beta-glucuronidase toward triterpenoid saponins.
J.Biol.Chem., 293, 2018
|
|
2CHL
 
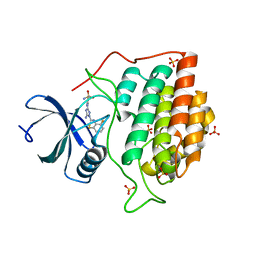 | | Structure of casein kinase 1 gamma 3 | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, CASEIN KINASE I ISOFORM GAMMA-3, SULFATE ION | | Authors: | Bunkoczi, G, Salah, E, Rellos, P, Das, S, Fedorov, O, Savitsky, P, Gileadi, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Ugochukwu, E, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-03-15 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Casein Kinase 1 Gamma 3
To be Published
|
|
5NNF
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated BAZ1B peptide (K221ac) | | Descriptor: | Bromodomain-containing protein 4, FLPH(ALY)YDVKL | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
8C14
 
 | | Aurora A kinase in complex with TPX2-inhibitor 9 | | Descriptor: | (1~{R},2~{R})-cyclohexane-1,2-dicarboxylic acid, 4-(4-chloranyl-3-cyano-phenyl)-1~{H}-indole-6-carboxylic acid, ACETATE ION, ... | | Authors: | Fischer, G, Rocaboy, M, Blaszczyk, B, Moschetti, T, Wang, X, Scott, D.E, Coyne, A.G, Dagostin, C, Rooney, T, Bayly, A, Feng, J, Asteian, A, Alcaide-Lopez, A, Stockwell, S, Skidmore, J, Venkitaraman, A.R, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Selective Aurora A-TPX2 Interaction Inhibitors Have In Vivo Efficacy as Targeted Antimitotic Agents.
J.Med.Chem., 67, 2024
|
|
2CE6
 
 | | Structure of Helix Pomatia agglutinin with no ligands | | Descriptor: | HELIX POMATIA AGGLUTININ | | Authors: | Sanchez, J.-F, Lescar, J, Chazalet, V, Audfray, A, Gautier, C, Gagnon, J, Breton, C, Imberty, A, Mitchell, E.P. | | Deposit date: | 2006-02-03 | | Release date: | 2006-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and Structural Analysis of Helix Pomatia Agglutinin. A Hexameric Lectin with a Novel Fold.
J.Biol.Chem., 281, 2006
|
|
5NNO
 
 | | Structure of TbALDH3 complexed with NAD and AN3057 aldehyde | | Descriptor: | 4-[(1-oxidanyl-3~{H}-2,1-benzoxaborol-5-yl)oxy]benzaldehyde, Aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zoltner, M, Zhang, N, Horn, D, Field, M.C. | | Deposit date: | 2017-04-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Host-parasite co-metabolic activation of antitrypanosomal aminomethyl-benzoxaboroles.
PLoS Pathog., 14, 2018
|
|
8C1D
 
 | | Aurora A kinase in complex with TPX2-inhibitor 9 | | Descriptor: | (1~{R},2~{R})-cyclohexane-1,2-dicarboxylic acid, 4-(4-chlorophenyl)-7-methyl-1~{H}-indole-6-carboxylic acid, ACETATE ION, ... | | Authors: | Fischer, G, Rocaboy, M, Blaszczyk, B, Moschetti, T, Wang, X, Scott, D.E, Coyne, A.G, Dagostin, C, Rooney, T, Bayly, A, Feng, J, Asteian, A, Alcaide-Lopez, A, Stockwell, S, Skidmore, J, Venkitaraman, A.R, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.115 Å) | | Cite: | Selective Aurora A-TPX2 Interaction Inhibitors Have In Vivo Efficacy as Targeted Antimitotic Agents.
J.Med.Chem., 67, 2024
|
|
