3N33
 
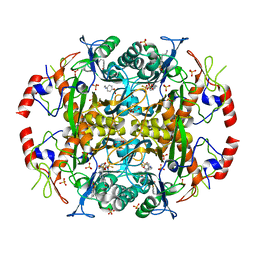 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with pefabloc SC (AEBSF) | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-peptidyl aminopeptidase, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P, Gruetter, M.G. | | Deposit date: | 2010-05-19 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Autoproteolytic and catalytic mechanisms for the beta-aminopeptidase BapA--a member of the Ntn hydrolase family.
Structure, 20, 2012
|
|
6DCB
 
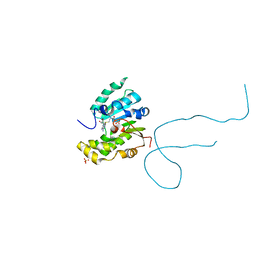 | | Structure of methylphosphate capping enzyme methyltransferase domain in complex with 5' end of 7SK RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yang, Y, Eichhorn, C, Cascio, D, Feigon, J. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis of 7SK RNA 5'-gamma-phosphate methylation and retention by MePCE.
Nat. Chem. Biol., 15, 2019
|
|
3VRT
 
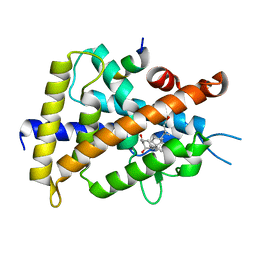 | | VDR ligand binding domain in complex with 2-Mehylidene-19,25,26,27-tetranor-1alpha,24-dihydroxyvitaminD3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-hydroxypentan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yoshimoto, N, Inaba, Y, Itoh, T, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
3VS2
 
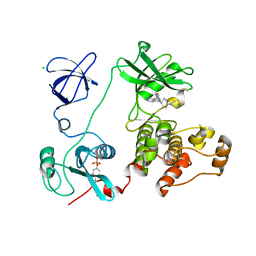 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[cis-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[cis-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Niwa, H, Parker, J.L, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
8PDI
 
 | | The phosphatase and C2 domains of SHIP1 with covalent Z1763271112 | | Descriptor: | (5-phenyl-1,3,4-thiadiazol-2-yl)methanimine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Moreira, T, Pascoa, T.C, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
5FFV
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with H3K14ac histone peptide | | Descriptor: | 1,2-ETHANEDIOL, Histone H3, Peregrin | | Authors: | Tallant, C, Nunez-Alonso, G, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2015-12-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the bromodomain of human BRPF1 in complex with H3K14ac histone peptide
To Be Published
|
|
6SI1
 
 | | p53 cancer mutant Y220H | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellular tumor antigen p53, ... | | Authors: | Joerger, A.C, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
8Q1S
 
 | | Pathogenic mutations of human phosphorylation sites affect protein-protein interactions | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, BROMIDE ION, ... | | Authors: | Roske, Y, Daumke, O, Rrustemi, T, Selbach, M. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Pathogenic mutations of human phosphorylation sites affect protein-protein interactions.
Nat Commun, 15, 2024
|
|
8PQP
 
 | |
1LZB
 
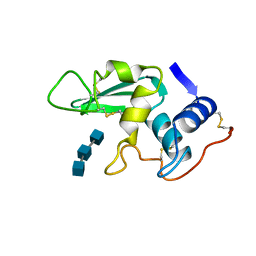 | | DISSECTION OF PROTEIN-CARBOHYDRATE INTERACTIONS IN MUTANT HEN EGG-WHITE LYSOZYME COMPLEXES AND THEIR HYDROLYTIC ACTIVITY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEN EGG WHITE LYSOZYME | | Authors: | Maenaka, K, Matsushima, M, Song, H, Watanabe, K, Kumagai, I. | | Deposit date: | 1995-02-10 | | Release date: | 1995-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dissection of protein-carbohydrate interactions in mutant hen egg-white lysozyme complexes and their hydrolytic activity.
J.Mol.Biol., 247, 1995
|
|
9GHN
 
 | | Structure of SARS-CoV-2 Main Protease (Mpro) with mutation of Q256A | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Creon, A, Scheer, T.E.S, Lane, T.J, Rahmani Mashhour, A, Guenther, S, Reinke, P.Y.A, Meents, A, Chapman, H.N. | | Deposit date: | 2024-08-15 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of SARS-CoV-2 Main Protease (Mpro) with mutation of Q256A
To Be Published
|
|
2P7Q
 
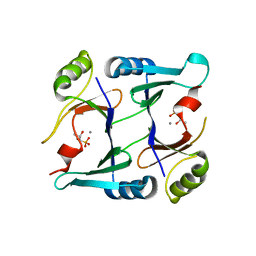 | | Crystal structure of E126Q mutant of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes complexed with MN(II) and 1S,2S-dihydroxypropylphosphonic acid | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION, [(1S,2S)-1,2-DIHYDROXYPROPYL]PHOSPHONIC ACID | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
5LQQ
 
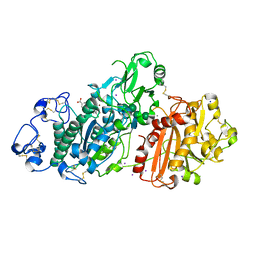 | | Structure of Autotaxin (ENPP2) with LM350 | | Descriptor: | 3-(6-chloranyl-2-methyl-1-phenyl-indol-3-yl)sulfanylbenzoic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Keune, W.J, Heidebrecht, T, Castelmur, E, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-08-17 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Activity Relationships of Small Molecule Autotaxin Inhibitors with a Discrete Binding Mode.
J. Med. Chem., 60, 2017
|
|
3VM7
 
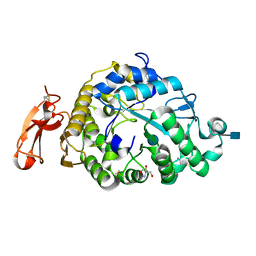 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | Deposit date: | 2011-12-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
4NT0
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 3-deazauridine 5'-monophosphate | | Descriptor: | 4-hydroxy-1-(5-O-phosphono-beta-D-ribofuranosyl)pyridin-2(1H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-11-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 3-deazauridine 5'-monophosphate
To be Published
|
|
8PJ8
 
 | | FKBP51FK1 F67E/K60Orn (i, i+7) in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Charalampidou, A, Hausch, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
3EFG
 
 | | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, Protein slyX homolog | | Authors: | Cuff, M.E, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913
TO BE PUBLISHED
|
|
9GOO
 
 | |
3MUD
 
 | | Structure of the Tropomyosin Overlap Complex from Chicken Smooth Muscle | | Descriptor: | 1,2-ETHANEDIOL, DNA repair protein XRCC4,Tropomyosin alpha-1 chain, SULFATE ION, ... | | Authors: | Klenchin, V.A, Frye, J, Rayment, I. | | Deposit date: | 2010-05-02 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition .
Biochemistry, 49, 2010
|
|
8PQV
 
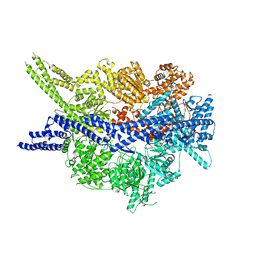 | | Cytoplasmic dynein-1 motor domain in post-powerstroke state | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1 | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
3E8R
 
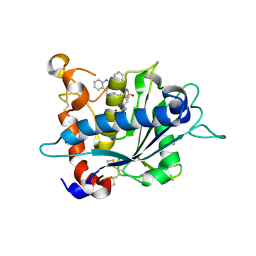 | | Crystal structure of catalytic domain of TACE with hydroxamate inhibitor | | Descriptor: | (1R,2S)-N~2~-hydroxy-1-{4-[(2-phenylquinolin-4-yl)methoxy]benzyl}cyclopropane-1,2-dicarboxamide, ADAM 17, CITRIC ACID, ... | | Authors: | Orth, P. | | Deposit date: | 2008-08-20 | | Release date: | 2008-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel hydroxamates as highly potent tumor necrosis factor-alpha converting enzyme inhibitors. Part II: optimization of the S3' pocket.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2P18
 
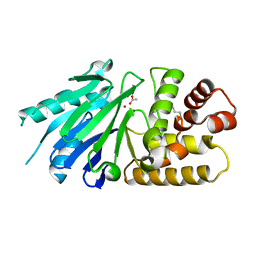 | | Crystal structure of the Leishmania infantum glyoxalase II | | Descriptor: | ACETIC ACID, Glyoxalase II, SPERMIDINE, ... | | Authors: | Trincao, J, Barata, L, Najmudin, S, Bonifacio, C, Romao, M.J. | | Deposit date: | 2007-03-02 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalysis and Structural Properties of Leishmania infantum Glyoxalase II: Trypanothione Specificity and Phylogeny.
Biochemistry, 47, 2008
|
|
8PFL
 
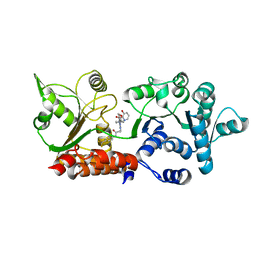 | | Crystal structure of WRN helicase domain in complex with 3 | | Descriptor: | 2-[2-(3,6-dihydro-2~{H}-pyran-4-yl)-5-ethyl-7-oxidanylidene-6-[4-(3-oxidanylpyridin-2-yl)carbonylpiperazin-1-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-4-yl]-~{N}-[2-methyl-4-(trifluoromethyl)phenyl]ethanamide, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION | | Authors: | Scheufler, C, Meyer, M, Moebitz, H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.
Nature, 629, 2024
|
|
2P2R
 
 | | Crystal structure of the third KH domain of human Poly(C)-Binding Protein-2 in complex with C-rich strand of human telomeric DNA | | Descriptor: | 6-AMINOPYRIMIDIN-2(1H)-ONE, C-rich strand of human telomeric DNA, Poly(rC)-binding protein 2 | | Authors: | James, T.L, Stroud, R.M, Du, Z, Fenn, S, Tjhen, R, Lee, J.K. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the third KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.6 A resolution.
Nucleic Acids Res., 35, 2007
|
|
3MY1
 
 | | Structure of CDK9/cyclinT1 in complex with DRB | | Descriptor: | 5,6-dichloro-1-beta-D-ribofuranosyl-1H-benzimidazole, Cell division protein kinase 9, Cyclin-T1, ... | | Authors: | Baumli, S, Johnson, L.N. | | Deposit date: | 2010-05-09 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Halogen bonds form the basis for selective P-TEFb inhibition by DRB
Chem.Biol., 17, 2010
|
|
