8SRQ
 
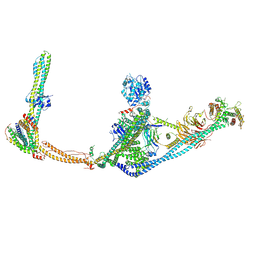 | | Structure of human ULK1C:PI3KC3-C1 supercomplex | | Descriptor: | Autophagy-related protein 13, Beclin 1-associated autophagy-related key regulator, Beclin-1-C 35 kDa, ... | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2023-05-05 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
8SRP
 
 | |
8SRO
 
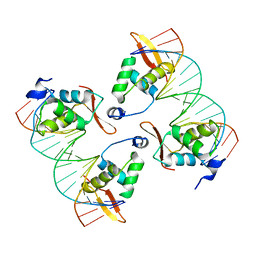 | | FoxP3 tetramer on TTTG repeats | | Descriptor: | DNA 72-mer, Forkhead box protein P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | FOXP3 recognizes microsatellites and bridges DNA through multimerization.
Nature, 624, 2023
|
|
8SRM
 
 | |
8SRL
 
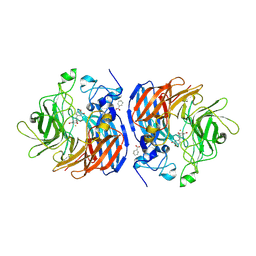 | |
8SRK
 
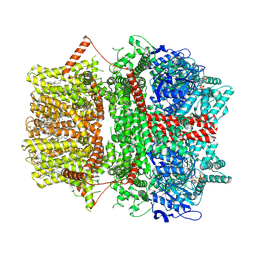 | | Cryo-EM structure of TRPM2 chanzyme (without NUDT9-H domain) in the presence of Ca and ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRJ
 
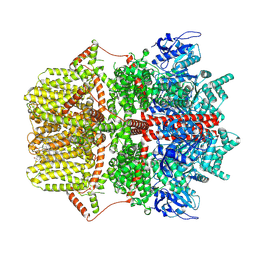 | |
8SRI
 
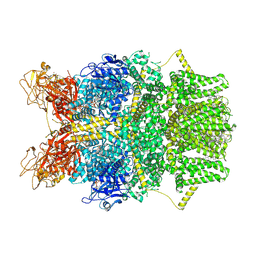 | | Cryo-EM structure of TRPM2 chanzyme (E1114A) in the presence of Magnesium and ADP-ribose, closed state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRH
 
 | | Cryo-EM structure of TRPM2 chanzyme (E1114A) in the presence of Magnesium and ADP-ribose, open state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRG
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, Adenosine monophosphate, and Ribose-5-phosphate | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRF
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, ADP-ribose, Adenosine monophosphate, and Ribose-5-phosphate, closed state | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRE
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium and ADP-ribose, closed state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRD
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium and ADP-ribose, open state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRC
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Calcium and ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
8SRB
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of EDTA and ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
8SRA
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Calcium | | Descriptor: | CALCIUM ION, CHOLESTEROL, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
8SR9
 
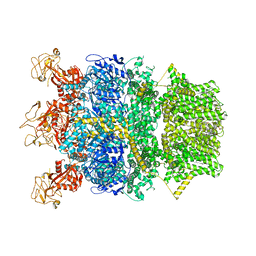 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
8SR8
 
 | |
8SR7
 
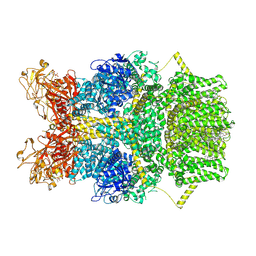 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, Adenosine monophosphate, and Ribose-5-phosphate | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.97 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
8SR6
 
 | |
8SR5
 
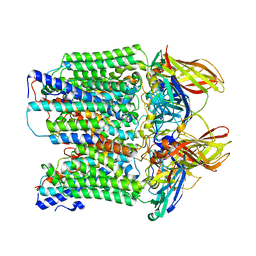 | |
8SR4
 
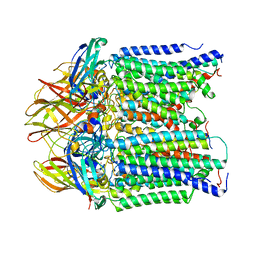 | | particulate methane monooxygeanse treated with potassium cyanide and copper reloaded | | Descriptor: | Ammonia monooxygenase/methane monooxygenase, subunit C family protein, COPPER (II) ION, ... | | Authors: | Tucci, F.J, Jodts, R.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8SR2
 
 | | particulate methane monooxygenase incubated with 4,4,4-trifluorobutanol | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Product analogue binding identifies the copper active site of particulate methane monooyxgenase
Nat Catal, 2023
|
|
8SR1
 
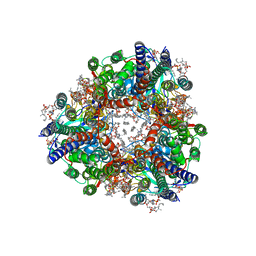 | | particulate methane monooxygenase crosslinked with 4,4,4-trifluorobutanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, 4,4,4-trifluorobutan-1-ol, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8SR0
 
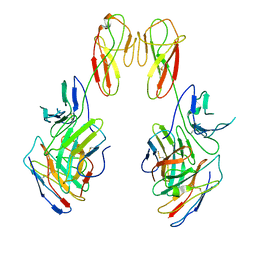 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 local refined | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
