9EMU
 
 | | RosC-8-demethyl-8-amino-FMN - Phosphate complex | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ermler, U, Mack, M, Demmer, U. | | Deposit date: | 2024-03-11 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Phosphatase RosC from Streptomyces davaonensis is Used for Roseoflavin Biosynthesis and has Evolved to Largely Prevent Dephosphorylation of the Important Cofactor Riboflavin-5'-phosphate.
J.Mol.Biol., 436, 2024
|
|
1N0H
 
 | | Crystal Structure of Yeast Acetohydroxyacid Synthase in Complex with a Sulfonylurea Herbicide, Chlorimuron Ethyl | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, 4-{[(4'-AMINO-2'-METHYLPYRIMIDIN-5'-YL)METHYL]AMINO}PENT-3-ENYL DIPHOSPHATE, ... | | Authors: | Pang, S.S, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2002-10-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of sulfonylurea herbicide inhibition of acetohydroxyacid synthase
J.BIOL.CHEM., 278, 2003
|
|
9EOY
 
 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CaMKII alpha hub bound to PIPA | | Descriptor: | 2-[6-(4-chlorophenyl)imidazo[1,2-b]pyridazin-2-yl]ethanoic acid, ACETATE ION, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Narayanan, D, Larsen, A.S.G, Solbak, S.M.O, Wellendorph, P, Gee, C.L, Kastrup, J.S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced CaMKII alpha hub Trp403 flip, hub domain stacking, and modulation of kinase activity.
Protein Sci., 33, 2024
|
|
5J25
 
 | | Endothiapepsin in complex with fragment 158 | | Descriptor: | 2-(4-methylpiperidin-1-yl)-N-[3-(propan-2-yl)-1,2-oxazol-5-yl]acetamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Z1V
 
 | | Structure of Factor Inhibiting HIF (FIH) in complex with Fe, NO, and NOG | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Taabazuing, C.Y, Garman, S.C, Knapp, M.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Promotes Productive Gas Binding in the alpha-Ketoglutarate-Dependent Oxygenase FIH.
Biochemistry, 55, 2016
|
|
8XR5
 
 | | Crystal structure of PD-L1 complexed with small molecule inhibitor X18 | | Descriptor: | (2~{R})-2-[[2-(2,1,3-benzoxadiazol-5-ylmethoxy)-5-chloranyl-4-[[2-fluoranyl-3-[3-[3-(4-oxidanylpiperidin-1-yl)propoxy]phenyl]phenyl]methoxy]phenyl]methylamino]-3-oxidanyl-propanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Liu, L. | | Deposit date: | 2024-01-06 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Novel PD-L1 Small-Molecular Inhibitors with Potent In Vivo Anti-tumor Immune Activity.
J.Med.Chem., 67, 2024
|
|
8X7H
 
 | | Crystal structure of the ternary complex of GID4-PROTAC(NEP162)-BRD4(BD1). | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, Glucose-induced degradation protein 4 homolog, ... | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2023-11-24 | | Release date: | 2025-03-26 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of PROTACs utilizing the E3 ligase GID4 for targeted protein degradation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5WIM
 
 | | JAK2 Pseudokinase in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
7UM9
 
 | | Human ALDH1A1 with bound compound CM38 | | Descriptor: | (4-methylfuro[3,2-c]quinolin-2-yl)(piperidin-1-yl)methanone, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hurley, T.D. | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel ALDH1A1 Inhibitor Blocks Platinum-Induced Senescence and Stemness in Ovarian Cancer.
Cancers (Basel), 14, 2022
|
|
6EI9
 
 | | Crystal structure of E. coli tRNA-dihydrouridine synthase B (DusB) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Bou-Nader, C, Hamdane, D. | | Deposit date: | 2017-09-18 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Unveiling structural and functional divergences of bacterial tRNA dihydrouridine synthases: perspectives on the evolution scenario.
Nucleic Acids Res., 46, 2018
|
|
8BTS
 
 | |
5J8I
 
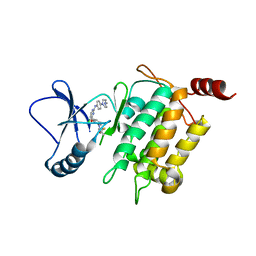 | | Crystal structure of TL11-113 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7/TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
9EHY
 
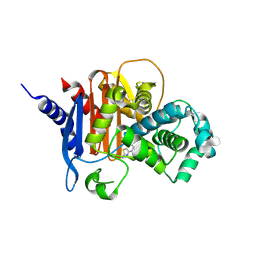 | | X-ray crystal structure of ADC-33 beta-lactamase in complex with ceftazidime in acyl and product forms | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Powers, R.A, Wallar, B.J, Jarvis, H.R, Ziegler, Z.X, June, C.M. | | Deposit date: | 2024-11-25 | | Release date: | 2025-05-14 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Resistance to oxyimino-cephalosporins conferred by an alternative mechanism of hydrolysis by the Acinetobacter -derived cephalosporinase-33 (ADC-33), a class C beta-lactamase present in carbapenem-resistant Acinetobacter baumannii (CR Ab ).
Mbio, 16, 2025
|
|
5L77
 
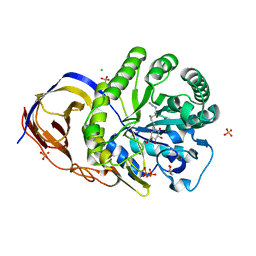 | | A glycoside hydrolase mutant with an unreacted activity based probe bound | | Descriptor: | (1~{R},2~{S},3~{R},4~{S},5~{S},6~{R})-7-[8-[(azanylidene-{4}-azanylidene)amino]octyl]-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-06-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
9EEI
 
 | |
4YFY
 
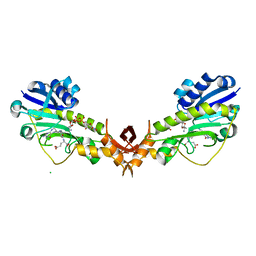 | | X-ray structure of the Viof N-formyltransferase from Providencia alcalifaciens O30 in complex with THF and TDP-Qui4N | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Genthe, N.A, Thoden, J.B, Benning, M.M, Holden, H.M. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structure of an N-formyltransferase from Providencia alcalifaciens O30.
Protein Sci., 24, 2015
|
|
5N9P
 
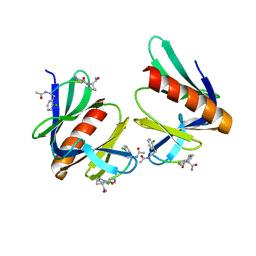 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-NH2 | | Descriptor: | Ac-[2-Cl-F]-PP-[ProM-1]-NH2, CHLORIDE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-26 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5J48
 
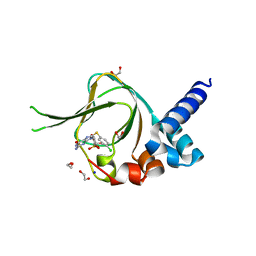 | | PKG I's Carboyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-pCPT-cGMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-8-[(4-chlorophenyl)sulfanyl]-9-[(2S,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-3,9-dihydro-6H-purin-6-one, CALCIUM ION, ... | | Authors: | Campbell, J.C, Sankaran, B, Kim, C.W. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Basis of Analog Specificity in PKG I and II.
ACS Chem. Biol., 12, 2017
|
|
9D1M
 
 | | Lucilia cuprina alpha esterase 7 directed evolution round 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Carboxylic ester hydrolase, ... | | Authors: | Frkic, R.L, Fraser, N, Mabbitt, P.D, Jackson, C.J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Lucilia cuprina alpha esterase 7 directed evolution round 2
To Be Published
|
|
9D1S
 
 | | Lucilia cuprina alpha esterase 7 directed evolution round 8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Frkic, R.L, Fraser, N, Mabbitt, P.D, Jackson, C.J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Lucilia cuprina alpha esterase 7 directed evolution round 8
To Be Published
|
|
6BNM
 
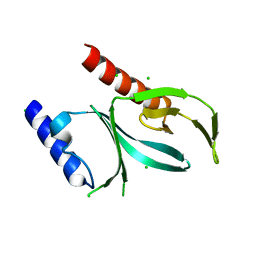 | | Crystal Structure of the P-Rex2 PH domain | | Descriptor: | CHLORIDE ION, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 2 protein | | Authors: | Cash, J.N, Sharma, P.V, Tesmer, J.J.G. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of the pleckstrin homology domain of the RhoGEF P-Rex2 and its regulation by PIP3.
J.Struct.Biol., 1, 2019
|
|
6FBH
 
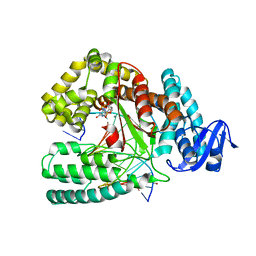 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the sixth primer nucleotide. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*TP*CP*CP*GP*GP*TP*GP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5J9U
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
6MG3
 
 | | V285A Mutant of the C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6MG2
 
 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2221 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
