6HN3
 
 | | wildtype form (apo) of human GPX4 with Se-Cys46 | | Descriptor: | CHLORIDE ION, ETHANOL, GLYCEROL, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Hoffmann, J, Schnirch, L, Eaton, J.K, Badock, V, Gradl, S. | | Deposit date: | 2018-09-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Crystal structures of the selenoprotein glutathione peroxidase 4 in its apo form and in complex with the covalently bound inhibitor ML162.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
9I17
 
 | | Human protein kinase CK2 alpha in complex with TN20 | | Descriptor: | (2~{Z},5~{Z})-2-(3-chlorophenyl)imino-5-[(4-methoxy-3-oxidanyl-phenyl)methylidene]-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Lolli, G. | | Deposit date: | 2025-01-16 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Switching off CK2-mediated activation of survivin offers new therapeutic opportunities in neuroblastoma
To Be Published
|
|
9I13
 
 | | Human protein kinase CK2 alpha in complex with TN19 | | Descriptor: | (2~{Z},5~{Z})-2-(3-fluorophenyl)imino-5-[(4-methoxy-3-oxidanyl-phenyl)methylidene]-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Lolli, G. | | Deposit date: | 2025-01-15 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Switching off CK2-mediated activation of survivin offers new therapeutic opportunities in neuroblastoma
To Be Published
|
|
5XBU
 
 | | Crystal structure of GH45 endoglucanase EG27II in apo-form | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
9I0Z
 
 | | Human protein kinase CK2 alpha in complex with TN11 | | Descriptor: | (2~{Z},5~{Z})-5-[(4-methoxy-3-oxidanyl-phenyl)methylidene]-2-(2-methylphenyl)imino-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Lolli, G. | | Deposit date: | 2025-01-15 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Switching off CK2-mediated activation of survivin offers new therapeutic opportunities in neuroblastoma
To Be Published
|
|
9I11
 
 | | Human protein kinase CK2 alpha in complex with TN16 | | Descriptor: | (2~{Z},5~{Z})-2-(3-hydroxyphenyl)imino-5-[(4-methoxy-3-oxidanyl-phenyl)methylidene]-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Lolli, G. | | Deposit date: | 2025-01-15 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Switching off CK2-mediated activation of survivin offers new therapeutic opportunities in neuroblastoma
To Be Published
|
|
9I10
 
 | | Human protein kinase CK2 alpha in complex with TN12 | | Descriptor: | (2~{Z},5~{Z})-5-[(4-methoxy-3-oxidanyl-phenyl)methylidene]-2-(3-methylphenyl)imino-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Lolli, G. | | Deposit date: | 2025-01-15 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Switching off CK2-mediated activation of survivin offers new therapeutic opportunities in neuroblastoma
To Be Published
|
|
9LG0
 
 | | Crystal Structure of Human Peroxiredoxin I in Complex with SAB | | Descriptor: | (2~{R})-3-[3,4-bis(oxidanyl)phenyl]-2-[(~{E})-3-[(2~{S},3~{S})-2-[3,4-bis(oxidanyl)phenyl]-3-[(2~{R})-3-[3,4-bis(oxidanyl)phenyl]-1-oxidanyl-1-oxidanylidene-propan-2-yl]oxycarbonyl-7-oxidanyl-2,3-dihydro-1-benzofuran-4-yl]prop-2-enoyl]oxy-propanoic acid, Peroxiredoxin-1 | | Authors: | Xu, H, Luo, C. | | Deposit date: | 2025-01-09 | | Release date: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Augmentation of PRDX1-DOK3 interaction alleviates rheumatoid arthritis progression by suppressing plasma cell differentiation.
Acta Pharm Sin B, 15, 2025
|
|
4R8V
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with formate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-09-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3I5M
 
 | | Structure of the apo form of leucoanthocyanidin reductase from vitis vinifera | | Descriptor: | Putative leucoanthocyanidin reductase 1 | | Authors: | Mauge, C, Gargouri, M, d'Estaintot, B.L, Granier, T, Gallois, B. | | Deposit date: | 2009-07-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure and catalytic mechanism of leucoanthocyanidin reductase from Vitis vinifera.
J.Mol.Biol., 397, 2010
|
|
3I6I
 
 | | Structure of the binary complex leucoanthocyanidin reductase - NADPH from vitis vinifera | | Descriptor: | ACETIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative leucoanthocyanidin reductase 1 | | Authors: | Mauge, C, Gargouri, M, d'Estaintot, B.L, Granier, T, Gallois, B. | | Deposit date: | 2009-07-07 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and catalytic mechanism of leucoanthocyanidin reductase from Vitis vinifera.
J.Mol.Biol., 397, 2010
|
|
8S9P
 
 | | 1:1:1 agrin/LRP4/MuSK complex | | Descriptor: | Agrin, Low-density lipoprotein receptor-related protein 4, Muscle, ... | | Authors: | Xie, T, Xu, G.J, Liu, Y, Quade, B, Lin, W.C, Bai, X.C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the assembly of the agrin/LRP4/MuSK signaling complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4Z2W
 
 | | Factor Inhibiting HIF in Complex with Fe, and Alpha-Ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Taabazuing, C.Y, Garman, S.C, Knapp, M.J. | | Deposit date: | 2015-03-30 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Promotes Productive Gas Binding in the alpha-Ketoglutarate-Dependent Oxygenase FIH.
Biochemistry, 55, 2016
|
|
5MWG
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ091 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[1,4-dimethyl-2,3-bis(oxidanylidene)-7-piperidin-1-yl-quinoxalin-6-yl]-5,6,7,8-tetrahydronaphthalene-2-sulfonamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
6HTI
 
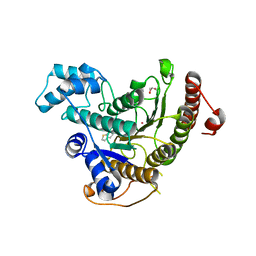 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 6 | | Descriptor: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
7CCP
 
 | |
6HEY
 
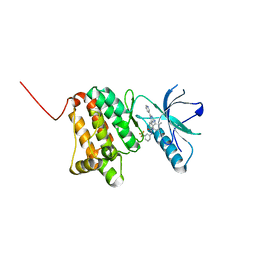 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative ATNK002 | | Descriptor: | 3-[(4,6-dipyridin-3-yl-1,3,5-triazin-2-yl)amino]-4-methyl-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.367 Å) | | Cite: | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
4QT0
 
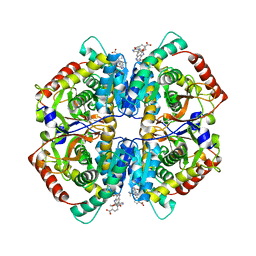 | | Crystal structure of human muscle L-lactate dehydrogenase in complex with inhibitor 1, 3-{[3-CARBAMOYL-7-(2,4-DIMETHOXYPYRIMIDIN-5-YL)QUINOLIN-4-YL]AMINO}BENZOIC ACID | | Descriptor: | 3-{[3-carbamoyl-7-(2,4-dimethoxypyrimidin-5-yl)quinolin-4-yl]amino}benzoic acid, L-lactate dehydrogenase A chain | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2014-07-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7C89
 
 | |
1TNL
 
 | |
9NR2
 
 | | Crystal structure of H5 hemagglutinin from the influenza virus A/black swan/Akita/1/2016 with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2025-03-13 | | Release date: | 2025-04-23 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Q226L mutation can convert a highly pathogenic H5 2.3.4.4e virus to bind human-type receptors.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
1MLD
 
 | | REFINED STRUCTURE OF MITOCHONDRIAL MALATE DEHYDROGENASE FROM PORCINE HEART AND THE CONSENSUS STRUCTURE FOR DICARBOXYLIC ACID OXIDOREDUCTASES | | Descriptor: | CITRIC ACID, MALATE DEHYDROGENASE | | Authors: | Gleason, W.B, Fu, Z, Birktoft, J.J, Banaszak, L.J. | | Deposit date: | 1994-01-24 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined crystal structure of mitochondrial malate dehydrogenase from porcine heart and the consensus structure for dicarboxylic acid oxidoreductases.
Biochemistry, 33, 1994
|
|
5MWZ
 
 | |
8DL9
 
 | |
5K2A
 
 | | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
