5LHI
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor N-[3-(ethoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]-4-methylthieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
9FMJ
 
 | | PsiM N247M in complex with sinefungin and baeocystin | | Descriptor: | Baeocystin, CHLORIDE ION, Psilocybin synthase, ... | | Authors: | Hudspeth, J, Rupp, B, Werten, S. | | Deposit date: | 2024-06-06 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The Second Methylation in Psilocybin Biosynthesis Is Enabled by a Hydrogen Bonding Network Extending into the Secondary Sphere Surrounding the Methyltransferase Active Site.
Chembiochem, 25, 2024
|
|
9BL0
 
 | | KRAS G12D Mutant KRAS 1-169 at 298 K bound to MRTX-1133 | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)-5-ethynyl-6-fluoronaphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Xu, M, Deck, S.L, Milano, S.K, Aplin, C, Cerione, R.A. | | Deposit date: | 2024-04-29 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Room Temperature Structures of Oncogenic KRAS-G12D bound to MRTX-1133
To Be Published
|
|
6G4N
 
 | | Torpedo californica acetylcholinesterase bound to uncharged hybrid reactivator 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[4-[(7-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]butyl]-2-[(oxidanylamino)methyl]pyridin-3-ol, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
6GE8
 
 | | Crystal structure of Mycobacterium tuberculosis BioA | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, MAGNESIUM ION, [(2~{S})-3-(1~{H}-indol-3-yl)-1-[(2~{E})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene]hydrazinyl]-1-oxidanylidene-propan-2-yl]azanium | | Authors: | Ekstrom, A, Campopiano, D.J, Marles-Wright, J. | | Deposit date: | 2018-04-25 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis BioA
To Be Published
|
|
6R4K
 
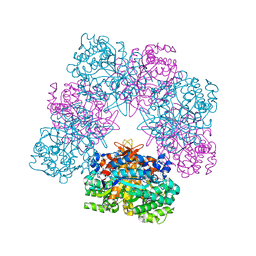 | | Structure of beta-glucosidase A from Paenibacillus polymyxa complexed with a monovalent inhibitor | | Descriptor: | (2~{S},3~{S},4~{R})-2-[[4-[4-[2-[2-(2-azanylidenehydrazinyl)ethoxy]ethoxy]phenyl]-1,2,3-triazol-1-yl]methyl]pyrrolidine-3,4-diol, Beta-glucosidase A | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis of the inhibition of GH1 beta-glucosidases by multivalent pyrrolidine iminosugars.
Bioorg.Chem., 89, 2019
|
|
8J79
 
 | |
5KT9
 
 | |
6BI2
 
 | |
6BHZ
 
 | | Trastuzumab Fab D185A (Light Chain) Mutant. | | Descriptor: | 1,2-ETHANEDIOL, Trastuzumab Anti-HER2 Fab Heavy Chain, Trastuzumab Anti-HER2 Fab Light Chain D185A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Chembiochem, 19, 2018
|
|
6HY7
 
 | |
5L6M
 
 | | Structure of Caulobacter crescentus VapBC1 (VapB1deltaC:VapC1 form) | | Descriptor: | GLYCEROL, MALONATE ION, Ribonuclease VapC, ... | | Authors: | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | Deposit date: | 2016-05-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
6K0J
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor | | Descriptor: | 3-(2,7-dimethoxyacridin-9-yl)sulfanylpropan-1-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Tiantian, W, Xiao, J. | | Deposit date: | 2019-05-06 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Inhibition of dual-specificity tyrosine phosphorylation-regulated kinase 2 perturbs 26S proteasome-addicted neoplastic progression.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HVF
 
 | | Kinase domain of cSrc in complex with compound 29B | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase Src, ~{N}-[3-[3-ethyl-6-[4-(4-methylpiperazin-1-yl)phenyl]-4-oxidanylidene-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]phenyl]prop-2-enamide | | Authors: | Keul, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of osimertinib-resistant epidermal growth factor receptor EGFR-T790M/C797S.
Chem Sci, 10, 2019
|
|
9G03
 
 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with ferredoxin (Clostridium autoethanogenum) | | Descriptor: | CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase/acetyl-CoA synthase beta subunit, Fe(3)-Ni(1)-S(4) cluster, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
9FZY
 
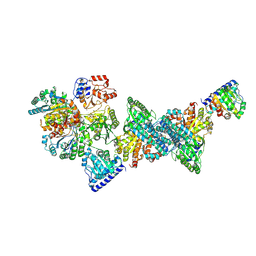 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with corrinoid iron-sulfur protein (CoFeSP) from Clostridium autoethanogenum (composite structure, class 3A) | | Descriptor: | Acetyl-CoA decarbonylase/synthase complex subunit delta, CO-methylating acetyl-CoA synthase, COBALAMIN, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
9FZZ
 
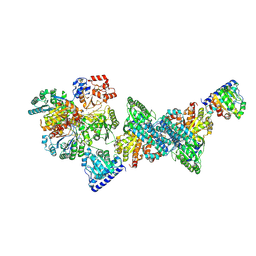 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with corrinoid iron-sulfur protein (CoFeSP) from Clostridium autoethanogenum (composite structure, class 3B) | | Descriptor: | Acetyl-CoA decarbonylase/synthase complex subunit delta, CO-methylating acetyl-CoA synthase, COBALAMIN, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
9G00
 
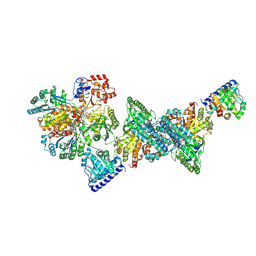 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) in complex with corrinoid iron-sulfur protein (CoFeSP) from Clostridium autoethanogenum (composite structure, class 3Cb) | | Descriptor: | Acetyl-CoA decarbonylase/synthase complex subunit delta, CO-methylating acetyl-CoA synthase, COBALAMIN, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
6SMY
 
 | |
9G02
 
 | | Structure of carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum (composite structure, semi-extended state) | | Descriptor: | CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase/acetyl-CoA synthase beta subunit, Fe(3)-Ni(1)-S(4) cluster, ... | | Authors: | Yin, M.D, Lemaire, O.N, Wagner, T, Murphy, B.J. | | Deposit date: | 2024-07-06 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Conformational dynamics of a multienzyme complex in anaerobic carbon fixation.
Science, 387, 2025
|
|
6GVC
 
 | | Structure of ArhGAP12 bound to G-Actin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Mouilleron, S, Treisman, R, Diring, J. | | Deposit date: | 2018-06-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RPEL-family rhoGAPs link Rac/Cdc42 GTP loading to G-actin availability.
Nat.Cell Biol., 21, 2019
|
|
7D2K
 
 | | Crystal structure of rat TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor Br-cis-22a | | Descriptor: | 1-(5-bromanylpyridin-3-yl)-4-[4-(3-methylphenyl)cyclohexyl]piperazin-4-ium, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Singh, A.K, Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.698 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
6I79
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 6-[(4-~{tert}-butyl-1,3-thiazol-2-yl)methyl]-4,6-diazaspiro[2.4]heptane-5,7-dione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6IMO
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[(1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6SPX
 
 | | Structure of protein kinase CK2 catalytic subunit in complex with the CK2beta-competitive bisubstrate inhibitor ARC1502 | | Descriptor: | 8-[4,5,6,7-tetrakis(bromanyl)benzimidazol-1-yl]octanoic acid, ARC1502, Casein kinase II subunit alpha | | Authors: | Niefind, K, Schnitzler, A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Unexpected CK2 beta-antagonistic functionality of bisubstrate inhibitors targeting protein kinase CK2.
Bioorg.Chem., 96, 2020
|
|
