7AQV
 
 | |
7YYK
 
 | | Crystal structure of the O-fucosylated form of TSRs1-3 from the human thrombospondin 1 | | Descriptor: | 1,2-ETHANEDIOL, Thrombospondin-1, alpha-L-fucopyranose | | Authors: | Berardinelli, S.J, Eletsky, A, Valero-Gonzalez, J, Ito, A, Manjunath, R, Hurtado-Guerrero, R, Prestegard, J.R, Woods, R.J, Haltiwanger, R.S. | | Deposit date: | 2022-02-18 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | O-fucosylation stabilizes the TSR3 motif in thrombospondin-1 by interacting with nearby amino acids and protecting a disulfide bond.
J.Biol.Chem., 298, 2022
|
|
5GU8
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) wild type | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain, SODIUM ION | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7YOW
 
 | |
4N12
 
 | | Crystal structure of human E18D DJ-1 in complex with Cu | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Protein DJ-1 | | Authors: | Cendron, L, Girotto, S, Bisaglia, M, Tessari, I, Mammi, S, Zanotti, G, Bubacco, L. | | Deposit date: | 2013-10-03 | | Release date: | 2014-03-05 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | DJ-1 Is a Copper Chaperone Acting on SOD1 Activation.
J.Biol.Chem., 289, 2014
|
|
5EYL
 
 | | TUBULIN-BINDING DARPIN | | Descriptor: | DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN), GLYCEROL, PHOSPHATE ION | | Authors: | Ahmad, S, Kossow, M, Gigant, B. | | Deposit date: | 2015-11-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Destabilizing an interacting motif strengthens the association of a designed ankyrin repeat protein with tubulin.
Sci Rep, 6, 2016
|
|
1QR4
 
 | | TWO FIBRONECTIN TYPE-III DOMAIN SEGMENT FROM CHICKEN TENASCIN | | Descriptor: | PROTEIN (TENASCIN) | | Authors: | Piontek, K, Bisig, D.A. | | Deposit date: | 1999-06-17 | | Release date: | 1999-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Purification, crystallization and preliminary crystallographic studies of a two fibronectin type-III domain segment from chicken tenascin encompassing the heparin- and contactin-binding regions.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6ZDT
 
 | | Crystal structure of eukaryotic Fibrillarin with Nop56 N-terminal domain | | Descriptor: | Nucleolar protein 56, rRNA 2'-O-methyltransferase fibrillarin | | Authors: | Hoefler, S, Lukat, P, Carlomagno, T, Blankenfeldt, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | High-resolution structure of eukaryotic Fibrillarin interacting with Nop56 amino-terminal domain.
Rna, 27, 2021
|
|
7Z1H
 
 | | VAR2CSA APO | | Descriptor: | VAR2CSA APO | | Authors: | Raghavan, S.S.R, Wang, K.T. | | Deposit date: | 2022-02-24 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM reveals the conformational epitope of human monoclonal antibody PAM1.4 broadly reacting with polymorphic malarial protein VAR2CSA.
Plos Pathog., 18, 2022
|
|
7Z12
 
 | | VAR2 complex with PAM1.4 | | Descriptor: | PAM1.4, Heavy Chain, light Chain, ... | | Authors: | Raghavan, S.S.R, Wang, K.T. | | Deposit date: | 2022-02-24 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals the conformational epitope of human monoclonal antibody PAM1.4 broadly reacting with polymorphic malarial protein VAR2CSA.
Plos Pathog., 18, 2022
|
|
5B4L
 
 | | Crystal structure of the catalytic domain of human PDE10A complexed with 1-(cyclopropylmethyl)-5-(2-(2,3-dihydro-1H-imidazo[1,2-a]benzimidazol-1-yl)ethoxy)-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one | | Descriptor: | 1-(cyclopropylmethyl)-5-(2-(2,3-dihydro-1H-imidazo[1,2-a]benzimidazol-1-yl)ethoxy)-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent and selective pyridazin-4(1H)-one-based PDE10A inhibitors interacting with Tyr683 in the PDE10A selectivity pocket
Bioorg.Med.Chem., 24, 2016
|
|
8B2T
 
 | | SARS-CoV-2 Main Protease (Mpro) in complex with nirmatrelvir alkyne | | Descriptor: | 3C-like proteinase nsp5, Nirmatrelvir (reacted form) | | Authors: | Owen, C.D, Crawshaw, A.D, Warren, A.J, Trincao, J, Zhao, Y, Brewitz, L, Malla, T.R, Salah, E, Petra, L, Strain-Damerell, C, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine.
J.Med.Chem., 66, 2023
|
|
5B4K
 
 | | Crystal structure of the catalytic domain of human PDE10A complexed with N-(4-((5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy)phenyl)-1H-benzimidazol-2-amine | | Descriptor: | MAGNESIUM ION, N-(4-((5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy)phenyl)-1H-benzimidazol-2-amine, ZINC ION, ... | | Authors: | Oki, H, Zama, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design and synthesis of potent and selective pyridazin-4(1H)-one-based PDE10A inhibitors interacting with Tyr683 in the PDE10A selectivity pocket
Bioorg.Med.Chem., 24, 2016
|
|
5DZN
 
 | | human T-cell immunoglobulin and mucin domain protein 4 | | Descriptor: | T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Yuan, S, Rao, Z, Wang, X. | | Deposit date: | 2015-09-25 | | Release date: | 2015-11-25 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIM-1 acts a dual-attachment receptor for Ebolavirus by interacting directly with viral GP and the PS on the viral envelope.
Protein Cell, 6, 2015
|
|
2KT4
 
 | | Lipocalin Q83 is a Siderocalin | | Descriptor: | Extracellular fatty acid-binding protein, GALLIUM (III) ION, N,N',N''-[(3S,7S,11S)-2,6,10-trioxo-1,5,9-trioxacyclododecane-3,7,11-triyl]tris(2,3-dihydroxybenzamide) | | Authors: | Coudevylle, N, Geist, L, Hartl, M, Kontaxis, G, Bister, K, Konrat, R. | | Deposit date: | 2010-01-18 | | Release date: | 2010-09-08 | | Last modified: | 2016-01-27 | | Method: | SOLUTION NMR | | Cite: | The v-myc-induced Q83 lipocalin is a siderocalin.
J.Biol.Chem., 285, 2010
|
|
4N0M
 
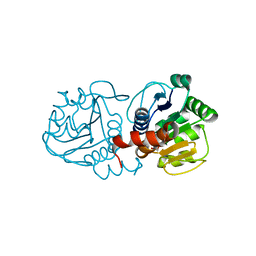 | | Crystal structure of human C53A DJ-1 in complex with Cu | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Protein DJ-1 | | Authors: | Cendron, L, Girotto, S, Bisaglia, M, Tessari, I, Mammi, S, Zanotti, G, Bubacco, L. | | Deposit date: | 2013-10-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DJ-1 Is a Copper Chaperone Acting on SOD1 Activation.
J.Biol.Chem., 289, 2014
|
|
5E1L
 
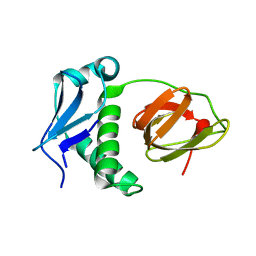 | | Structural and functional analysis of the E. coli FtsZ interacting protein, ZapC, reveals insight into molecular properties of a novel Z ring stabilizing protein | | Descriptor: | Cell division protein ZapC | | Authors: | Schumacher, M.A, Huang, K.-H, Tchorzewski, L, Zeng, W, Janakiraman, A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Analyses Reveal Insights into the Molecular Properties of the Escherichia coli Z Ring Stabilizing Protein, ZapC.
J.Biol.Chem., 291, 2016
|
|
2F20
 
 | | X-ray Crystal Structure of Protein BT_1218 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR8. | | Descriptor: | conserved hypothetical protein, with conserved domain | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Forouhar, F, Xiao, R, Ma, L.-C, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-11-15 | | Release date: | 2005-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Q8A8E9_BACTIN hypothetical protein from Bacteroides thetaiotaomicron.
To be Published
|
|
4MTC
 
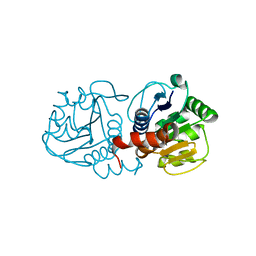 | | Crystal structure of human C53A DJ-1 | | Descriptor: | 1,2-ETHANEDIOL, Protein DJ-1 | | Authors: | Cendron, L, Girotto, S, Bisaglia, M, Tessari, I, Mammi, S, Zanotti, G, Bubacco, L. | | Deposit date: | 2013-09-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | DJ-1 Is a Copper Chaperone Acting on SOD1 Activation.
J.Biol.Chem., 289, 2014
|
|
8GBE
 
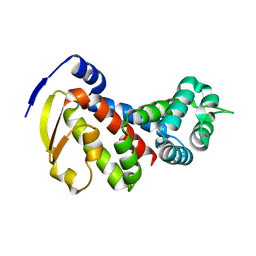 | |
8GIU
 
 | | Mycobacterium phage Patience | | Descriptor: | Capsid protein, gp_22 (Minor Capsid Protein), gp_4 (capsid accessory protein) | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2023-03-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | A novel accessory protein stabilizes the capsid of two actinobacteriophages
To Be Published
|
|
8E4X
 
 | |
8E0F
 
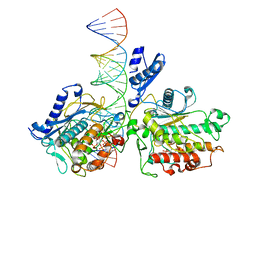 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2-RD) bound to dsRNA containing a G-G pair adjacent to the target site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5-R(*GP*CP*UP*CP*GP*CP*GP*AP*UP*GP*CP*GP*(8AZ)P*GP*AP*GP*GP*GP*CP* UP*CP*UP*GP*AP*UP*AP*GP*CP*UP*AP*CP*G)-3), ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ADAR activation by inducing a syn conformation at guanosine adjacent to an editing site.
Nucleic Acids Res., 50, 2022
|
|
2VSQ
 
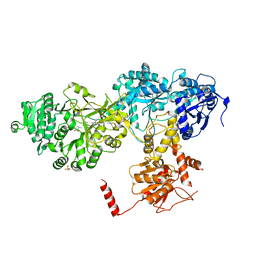 | | Structure of surfactin A synthetase C (SrfA-C), a nonribosomal peptide synthetase termination module | | Descriptor: | LEUCINE, SULFATE ION, SURFACTIN SYNTHETASE SUBUNIT 3 | | Authors: | Tanovic, A, Samel, S.A, Essen, L.-O, Marahiel, M.A. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the termination module of a nonribosomal peptide synthetase.
Science, 321, 2008
|
|
3K3K
 
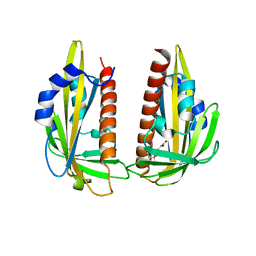 | | Crystal structure of dimeric abscisic acid (ABA) receptor pyrabactin resistance 1 (PYR1) with ABA-bound closed-lid and ABA-free open-lid subunits | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism of abscisic acid binding and signaling by dimeric PYR1.
Science, 326, 2009
|
|
