3B67
 
 | |
1Y95
 
 | | HIV-1 Dis(Mal) Duplex Pb-Soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', LEAD (II) ION, MAGNESIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
3BEJ
 
 | | Structure of human FXR in complex with MFA-1 and co-activator peptide | | Descriptor: | (8alpha,10alpha,13alpha,17beta)-17-[(4-hydroxyphenyl)carbonyl]androsta-3,5-diene-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Soisson, S.M, Parthasarathy, G, Becker, J.W. | | Deposit date: | 2007-11-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a potent synthetic FXR agonist with an unexpected mode of binding and activation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZW7
 
 | |
4QTQ
 
 | |
1Y90
 
 | | HIV-1 Dis(Mal) Duplex Mn-Soaked | | Descriptor: | 5'-R(*CP*(5BU)P*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MANGANESE (II) ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
6ZLX
 
 | | The structure of the ClpX-associated factor PDIP38 | | Descriptor: | PLATINUM (II) ION, Polymerase delta-interacting protein 2 | | Authors: | Zeth, K, Dougan, D. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | PDIP38 is a novel adaptor-like modulator of the mitochondrial AAA+ protease CLPXP
To Be Published
|
|
2ZW5
 
 | |
6ZWK
 
 | | Crystal structure of the phosphorylated C-terminal tail of histone H2AX in complex with a specific nanobody (C6 gammaXbody) | | Descriptor: | CHLORIDE ION, Histone H2AX, SODIUM ION, ... | | Authors: | McEwen, A.G, Moeglin, E, Desplancq, D, Weiss, E, Poterszman, A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Novel Nanobody Precisely Visualizes Phosphorylated Histone H2AX in Living Cancer Cells under Drug-Induced Replication Stress.
Cancers (Basel), 13, 2021
|
|
4P43
 
 | |
3C3K
 
 | | Crystal structure of an uncharacterized protein from Actinobacillus succinogenes | | Descriptor: | Alanine racemase, CHLORIDE ION, GLYCEROL | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of an uncharacterized protein from Actinobacillus succinogenes.
To be Published
|
|
4TK5
 
 | | Crystal Structure of human Tankyrase 2 in complex with EB47. | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TMK
 
 | |
1YCO
 
 | | Crystal structure of a branched-chain phosphotransacylase from Enterococcus faecalis V583 | | Descriptor: | PHOSPHATE ION, branched-chain phosphotransacylase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-22 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a branched-chain phosphotransacylase from Enterococcus faecalis V583
To be Published
|
|
2ZBS
 
 | |
1Y73
 
 | | HIV-1 Dis(Mal) Duplex Pt-Soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, PLATINUM (IV) ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
1Y23
 
 | |
7DSU
 
 | | Structure of Mod subunit of the Type III restriction-modification enzyme Mbo45V | | Descriptor: | 1,2-ETHANEDIOL, Mbo45V, SINEFUNGIN | | Authors: | Ahmed, I, Chouhan, O.P, Gopinath, A, Morgan, R.D, Bhagat, K, Singh, A, Saikrishnan, K. | | Deposit date: | 2021-01-02 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Mod subunit of the Type III restriction-modification enzyme Mbo45V
To Be Published
|
|
2ZAS
 
 | | Crystal structure of human estrogen-related receptor gamma ligand binding domain complex with 4-alpha-cumylphenol, a bisphenol A derivative | | Descriptor: | 4-(1-methyl-1-phenylethyl)phenol, Estrogen-related receptor gamma, GLYCEROL | | Authors: | Matsushima, A, Kakuta, Y, Teramoto, T, Shimohigashi, Y. | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ERRgamma tethers strongly bisphenol A and 4-alpha-cumylphenol in an induced-fit manner
Biochem.Biophys.Res.Commun., 373, 2008
|
|
4PML
 
 | | Crystal Structure of human Tankyrase 2 in complex with 3-amino-benzamide. | | Descriptor: | 1,2-ETHANEDIOL, 3-aminobenzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PNL
 
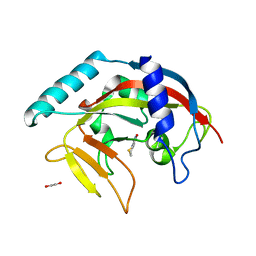 | | Crystal structure of TNKS-2 in complex with DR2313. | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,7,8-tetrahydro-4H-thiopyrano[4,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3BRQ
 
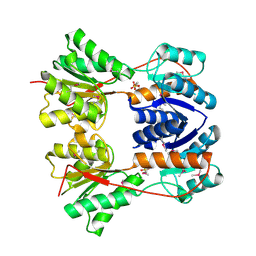 | | Crystal structure of the Escherichia coli transcriptional repressor ascG | | Descriptor: | HTH-type transcriptional regulator ascG, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Kagan, O, Evdokimova, E, Osipiuk, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the E. coli transcriptional repressor ascG.
To be Published
|
|
1ZB6
 
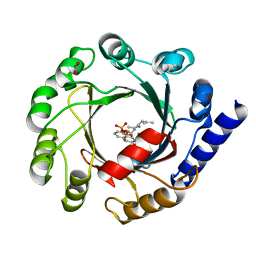 | | Co-Crystal Structure of ORF2 an Aromatic Prenyl Transferase from Streptomyces sp. strain cl190 complexed with GSPP and 1,6-dihydroxynaphtalene | | Descriptor: | 1,6-DIHYDROXY NAPHTHALENE, Aromatic prenyltransferase, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Kuzuyama, T, Noel, J.P, Richard, S.B. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products.
Nature, 435, 2005
|
|
1Z7S
 
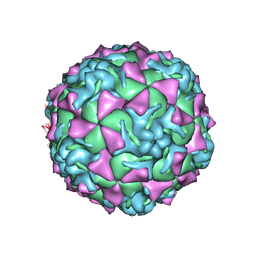 | | The crystal structure of coxsackievirus A21 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, Human COXSACKIEVIRUS A21, ... | | Authors: | Xiao, C, Bator-Kelly, C.M, Rieder, E, Chipman, P.R, Craig, A, Kuhn, R.J, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of coxsackievirus a21 and its interaction with icam-1.
Structure, 13, 2005
|
|
1ZMO
 
 | | Apo structure of haloalcohol dehalogenase HheA of Arthrobacter sp. AD2 | | Descriptor: | halohydrin dehalogenase | | Authors: | de Jong, R.M, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of the haloalcohol dehalogenase HheA from Arthrobacter sp. strain AD2: insight into enantioselectivity and halide binding in the haloalcohol dehalogenase family.
J.Bacteriol., 188, 2006
|
|
