5F8M
 
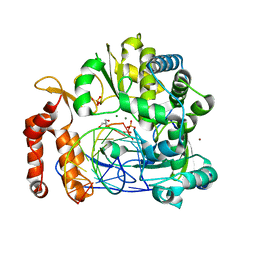 | | Enterovirus 71 Polymerase Elongation Complex (C3S4/5 Form) | | Descriptor: | GLYCEROL, Genome polyprotein, MAGNESIUM ION, ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2VT5
 
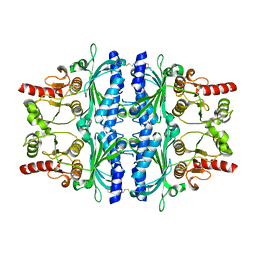 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE -1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH A DUAL BINDING AMP SITE INHIBITOR | | Descriptor: | 4-AMINO-N-[(2-SULFANYLETHYL)CARBAMOYL]BENZENESULFONAMIDE, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric Fbpase Inhibitors Gain 10(5) Times in Potency When Simultaneously Binding Two Neighboring AMP Sites.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8K71
 
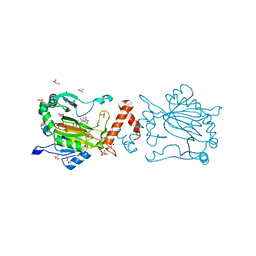 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((2-((naphthalen-2-ylmethyl)sulfonyl)acetyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | Descriptor: | 2-[(2~{Z})-2-[2-(naphthalen-2-ylmethylsulfonyl)ethanoylimino]-3-oxidanyl-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
2W97
 
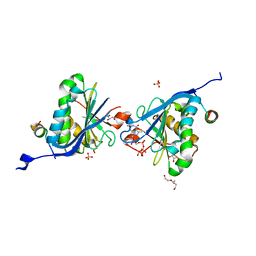 | | Crystal Structure of eIF4E Bound to Glycerol and eIF4G1 peptide | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4 GAMMA 1, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, GLYCEROL, ... | | Authors: | Brown, C.J, Verma, C.S, Walkinshaw, M.D, Lane, D.P. | | Deposit date: | 2009-01-22 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystallization of eIF4E complexed with eIF4GI peptide and glycerol reveals distinct structural differences around the cap-binding site.
Cell Cycle, 8, 2009
|
|
8K72
 
 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((3-(phenylsulfonamido)propanoyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | Descriptor: | 2-[(2~{Z})-3-oxidanyl-2-[3-(phenylsulfonylamino)propanoylimino]-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
2WA4
 
 | | FACTOR INHIBITING HIF-1 ALPHA WITH N,3-dihydroxybenzamide | | Descriptor: | FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N,3-DIHYDROXYBENZAMIDE, ... | | Authors: | Conejo-Garcia, A, Lienard, B.M.R, Clifton, I.J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2009-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
8K73
 
 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((1-(phenylsulfonyl)pyrrolidine-3-carbonyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | Descriptor: | 2-[(2~{Z})-3-oxidanyl-2-[(3~{R})-1-(phenylsulfonyl)pyrrolidin-3-yl]carbonylimino-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
6V8K
 
 | |
3S9S
 
 | | Ligand binding domain of PPARgamma complexed with a benzimidazole partial agonist | | Descriptor: | 1-(3,4-dichlorobenzyl)-2-methyl-N-[(1R)-1-phenylpropyl]-1H-benzimidazole-5-carboxamide, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Lambert, M.H, Xu, R.X. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of GSK1997132B a novel centrally penetrant benzimidazole PPARgamma partial agonist.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2VY6
 
 | | Two domains from the C-terminal region of influenza A virus polymerase PB2 subunit | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Crepin, T, Guilligay, D, Ruigrok, R, Cusack, S, Hart, D. | | Deposit date: | 2008-07-18 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Host Determinant Residue Lysine 627 Lies on the Surface of a Discrete, Folded Domain of Influenza Virus Polymerase Pb2 Subunit
Plos Pathog., 4, 2008
|
|
6SWS
 
 | | The DBB dimerization domain of B-cell adaptor for PI3K (BCAP) is required for down regulation of inflammatory signalling through the Toll-like receptor pathway | | Descriptor: | Phosphoinositide 3-kinase adapter protein 1 | | Authors: | Lauenstein, J.U, Scherm, M.J, Udgata, A, Moncrieffe, M.C, Fisher, D, Gay, N.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Negative Regulation of TLR Signaling by BCAP Requires Dimerization of Its DBB Domain.
J Immunol., 204, 2020
|
|
2VYT
 
 | | The MBT repeats of human SCML2 bind to peptides containing mono methylated lysine. | | Descriptor: | N-METHYL-LYSINE, SEX COMB ON MIDLEG-LIKE PROTEIN 2, TRIETHYLENE GLYCOL | | Authors: | Santiveri, C.M, Lechtenberg, B.C, Allen, M.D, Sathyamurthy, A, Jaulent, A.M, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2008-07-28 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Malignant Brain Tumor Repeats of Human Scml2 Bind to Peptides Containing Monomethylated Lysine.
J.Mol.Biol., 382, 2008
|
|
2W5Y
 
 | | Binary Complex of the Mixed Lineage Leukaemia (MLL1) SET Domain with the cofactor product S-Adenosylhomocysteine. | | Descriptor: | HISTONE-LYSINE N-METHYLTRANSFERASE HRX, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Southall, S.M, Wong, P.S, Odho, Z, Roe, S.M, WIlson, J.R. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Requirement of Additional Factors for Mll1 Set Domain Activity and Recognition of Epigenetic Marks.
Mol.Cell, 33, 2009
|
|
1KVA
 
 | | E. COLI RIBONUCLEASE HI D134A MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
6RBD
 
 | | State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
2WA3
 
 | | FACTOR INHIBITING HIF-1 ALPHA WITH 2-(3-hydroxyphenyl)-2-oxoacetic acid | | Descriptor: | 2-(3-HYDROXYPHENYL)-2-OXO-ETHANOIC ACID, FE (II) ION, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR | | Authors: | Conejo-Garcia, A, Lienard, B.M.R, Clifton, I.J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2009-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
6V2W
 
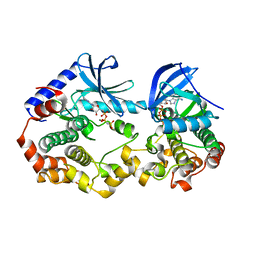 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Park, E, Eck, M.J. | | Deposit date: | 2019-11-25 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2WBD
 
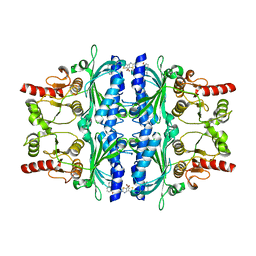 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE-1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH AN AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-3-ethylbenzenesulfonamide | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Kitas, E, Mohr, P, Kuhn, B, Wessel, H.P, Hebeisen, P, Haap, W, Huber, W, Alvarez Sanchez, R, Paehler, A, Bernadeau, A, Gubler, M, Schott, B, Tozzo, E. | | Deposit date: | 2009-02-26 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sulfonylureido Thiazoles as Fructose-1,6-Bisphosphatase Inhibitors for the Treatment of Type-2 Diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5KCE
 
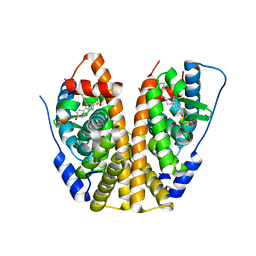 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl, 2-chlorobenzyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-(2-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-methyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
6V8N
 
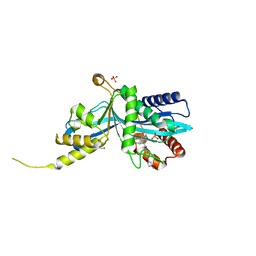 | |
6RBE
 
 | | State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
4R7P
 
 | | Human UDP-glucose pyrophosphorylase isoform 1 in complex with UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Fuehring, J, Cramer, J.T, Schneider, J, Baruch, P, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2014-08-28 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A Quaternary Mechanism Enables the Complex Biological Functions of Octameric Human UDP-glucose Pyrophosphorylase, a Key Enzyme in Cell Metabolism.
Sci Rep, 5
|
|
6GU3
 
 | | CDK1/CyclinB/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
8KDB
 
 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
4Y65
 
 | |
