3H61
 
 | | Catalytic domain of human Serine/Threonine Phosphatase 5 (PP5c) with two Mn2+ atoms originally soaked with norcantharidin (which is present in the structure in the hydrolyzed form) | | Descriptor: | (1R,2S,3R,4S)-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, MANGANESE (II) ION, Serine/threonine-protein phosphatase 5 | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Talluri, E. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis of serine/threonine phosphatase inhibition by the archetypal small molecules cantharidin and norcantharidin
J.Med.Chem., 52, 2009
|
|
2GX1
 
 | | Solution structure and alanine scan of a spider toxin that affects the activation of mammalian sodium channels | | Descriptor: | Neurotoxin magi-5 | | Authors: | Sabo, J.K, Corzo, G, Bosmans, F, Billen, B, Villegas, E, Tytgat, J, Norton, R.S. | | Deposit date: | 2006-05-08 | | Release date: | 2006-12-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Alanine Scan of a Spider Toxin That Affects the Activation of Mammalian Voltage-gated Sodium Channels
J.Biol.Chem., 282, 2007
|
|
1BMB
 
 | | GRB2-SH2 DOMAIN IN COMPLEX WITH KPFY*VNVEF (PKF270-974) | | Descriptor: | PROTEIN (GROWTH FACTOR RECEPTOR BOUND PROTEIN 2), PROTEIN (PKF270-974) | | Authors: | Rondeau, J.M, Zurini, M. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and conformational requirements for high-affinity binding to the SH2 domain of Grb2(1).
J.Med.Chem., 42, 1999
|
|
4FHI
 
 | | Development of synthetically accessible non-secosteroidal hybrid molecules combining vitamin D receptor agonism and histone deacetylase inhibition | | Descriptor: | N-hydroxy-2-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetamide, SRC-1, Vitamin Nuclear Receptor | | Authors: | Fischer, J, Wang, T.T, Kaldre, D, Rochel, N, Moras, D, White, J.H, Gleason, J.L. | | Deposit date: | 2012-06-06 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthetically accessible non-secosteroidal hybrid molecules combining vitamin d receptor agonism and histone deacetylase inhibition.
Chem.Biol., 19, 2012
|
|
1BM2
 
 | |
2MCG
 
 | |
3E8Z
 
 | |
3QC6
 
 | | GspB | | Descriptor: | CALCIUM ION, GLYCEROL, NITROGEN MOLECULE, ... | | Authors: | Pyburn, T.M. | | Deposit date: | 2011-01-15 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural Model for Binding of the Serine-Rich Repeat Adhesin GspB to Host Carbohydrate Receptors.
Plos Pathog., 7, 2011
|
|
2IYQ
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with shikimate and ADP | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-21 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IYS
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with shikimate, open LID (conf. A) | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-21 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IYY
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with shikimate-3-phosphate and SO4 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IVZ
 
 | | Structure of TolB in complex with a peptide of the colicin E9 T- domain | | Descriptor: | CALCIUM ION, COLICIN-E9, PROTEIN TOLB | | Authors: | Loftus, S.R, Walker, D, Mate, M.J, Bonsor, D.A, James, R, Moore, G.R, Kleanthous, C. | | Deposit date: | 2006-06-23 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Competitive Recruitment of the Periplasmic Translocation Portal Tolb by a Natively Disordered Domain of Colicin E9
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3VMP
 
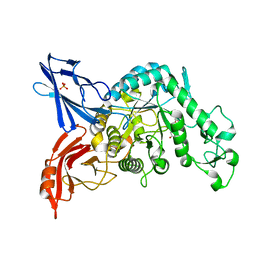 | | Crystal structure of dextranase from Streptococcus mutans in complex with 4,5-epoxypentyl alpha-D-glucopyranoside | | Descriptor: | 5-hydroxypentyl alpha-D-glucopyranoside, Dextranase, PHOSPHATE ION | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Okuyama, M, Mori, H, Funane, K, Kimura, A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural elucidation of dextran degradation mechanism by streptococcus mutans dextranase belonging to glycoside hydrolase family 66
J.Biol.Chem., 287, 2012
|
|
1SPU
 
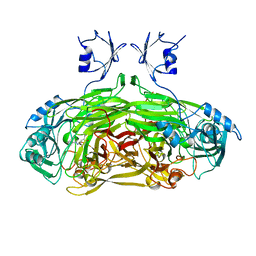 | | STRUCTURE OF OXIDOREDUCTASE | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Wilmot, C.M, Phillips, S.E.V. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction.
Biochemistry, 36, 1997
|
|
4G9Q
 
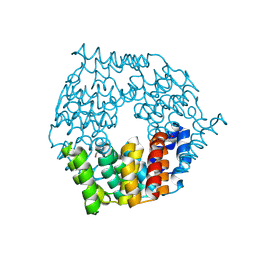 | | Crystal structure of a 4-carboxymuconolactone decarboxylase | | Descriptor: | 4-carboxymuconolactone decarboxylase | | Authors: | Hickey, H.D, Mcgillick, B.E, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-24 | | Release date: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a 4-carboxymuconolactone decarboxylase
To be Published
|
|
2MCC
 
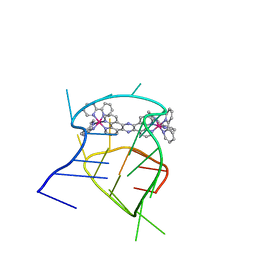 | | Structural studies on dinuclear ruthenium(II) complexes that bind diastereoselectively to an anti-parallel folded human telomere sequence | | Descriptor: | human_telomere_quadruplex, tetrakis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)(mu-tetrapyrido[3,2-a:2',3'-c:3'',2''-h:2''',3'''-j]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~13~,N~14~)diruthenium(4+) | | Authors: | Williamson, M.P, Wilson, T, Thomas, J.A, Felix, V, Costa, P.J. | | Deposit date: | 2013-08-18 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence.
J.Med.Chem., 56, 2013
|
|
3M4V
 
 | | Crystal structure of the A330P mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Whitehouse, C.J.C, Bell, S.G, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-03-12 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the properties of two single-site proline mutants of CYP102A1 (P450BM3)
Chembiochem, 11, 2010
|
|
3QH0
 
 | |
1XG4
 
 | | Crystal Structure of the C123S 2-Methylisocitrate Lyase Mutant from Escherichia coli in complex with the inhibitor isocitrate | | Descriptor: | ISOCITRIC ACID, MAGNESIUM ION, Probable methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Han, Y, Melamud, E, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of 2-Methylisocitrate Lyase in Complex with Product and with Isocitrate Inhibitor Provide Insight into Lyase Substrate Specificity, Catalysis and Evolution
Biochemistry, 44, 2005
|
|
2MYA
 
 | |
2MYC
 
 | |
1Z62
 
 | | Indirubin-3'-aminooxy-acetate inhibits glycogen phosphorylase by binding at the inhibitor and the allosteric site. Broad specificities of the two sites | | Descriptor: | ({[(3E)-2'-OXO-2',7'-DIHYDRO-2,3'-BIINDOL-3(7H)-YLIDENE]AMINO}OXY)ACETIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Kosmopoulou, M.N, Leonidas, D.D, Chrysina, E.D, Eisenbrand, G, Oikonomakos, N.G. | | Deposit date: | 2005-03-21 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Indirubin-3'-Aminooxy-Acetate Inhibits Glycogen Phosphorylase by Binding at the Inhibitor and the Allosteric Site. Broad Specificities of the Two Sites
LETT.DRUG DES.DISCOVERY, 2, 2005
|
|
1Z4Z
 
 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) with ligand DANA(soaked with sialic acid, pH7.0)) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
1ZB8
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
2JHD
 
 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 3'-sialyl-N-acetyllactosamine | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Blumenschein, T.M.A, Friedrich, N, Childs, R.A, Saouros, S, Carpenter, E.P, Campanero-Rhodes, M.A, Simpson, P, Koutroukides, T, Blackman, M.J, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2007-02-21 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Resolution Insight Into Host Cell Recognition by Toxoplasma Gondii.
Embo J., 26, 2007
|
|
