6MD1
 
 | |
1DF7
 
 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND METHOTREXATE | | Descriptor: | DIHYDROFOLATE REDUCTASE, GLYCEROL, METHOTREXATE, ... | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-17 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
1DFJ
 
 | |
1DG7
 
 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND 4-BROMO WR99210 | | Descriptor: | 1-[3-(4-BROMO-PHENOXY)-PROPOXY]-6,6-DIMETHYL-1.6-DIHYDRO-[1,3,5]TRIAZINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE, GLYCEROL, ... | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-23 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
6MCZ
 
 | |
7C4O
 
 | | Solution structure of the Orange domain from human protein HES1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Fan, J.S, Nayak, A, Swaminathan, K. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Induction of Transcriptional Inhibitor Hairy and Enhancer of Split Homolog-1 and the Related Repression of Tumor-Suppressor Thioredoxin-Interacting Protein Are Important Components of Cell-Transformation Program Imposed by Oncogenic Kinase Nucleophosmin-Anaplastic Lymphoma Kinase.
Am J Pathol, 2022
|
|
4Z3P
 
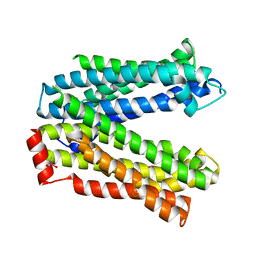 | | MATE transporter ClbM in complex with Rb+ | | Descriptor: | CACODYLATE ION, Putative drug/sodium antiporter, RUBIDIUM ION | | Authors: | Mousa, J.J, Bruner, S.D. | | Deposit date: | 2015-03-31 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | MATE transport of the E. coli-derived genotoxin colibactin.
Nat Microbiol, 1, 2016
|
|
6LXA
 
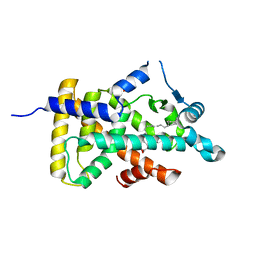 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
8P0A
 
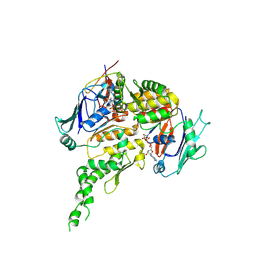 | | Human Cohesin ATPase module | | Descriptor: | 64-kDa C-terminal product, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Landwerlin, P, Durand, A, Diebold-Durand, M.-L, Romier, C. | | Deposit date: | 2023-05-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Human Cohesin ATPase module
To Be Published
|
|
1DGV
 
 | |
4Z3N
 
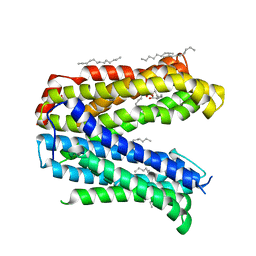 | | Crystal structure of the MATE transporter ClbM | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CACODYLATE ION, Putative drug/sodium antiporter | | Authors: | Mousa, J.J, Bruner, S.D. | | Deposit date: | 2015-03-31 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | MATE transport of the E. coli-derived genotoxin colibactin.
Nat Microbiol, 1, 2016
|
|
7WOX
 
 | | PPARgamma antagonist (MMT-160)- PPARgamma LBD complex | | Descriptor: | N-[[5-(3-phenylprop-2-ynoylamino)-2-propoxy-phenyl]methyl]-4-pyrimidin-2-yl-benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Yoshizawa, M, Aoyama, T, Itoh, T, Miyachi, H. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Arylalkynyl amide-type peroxisome proliferator-activated receptor gamma (PPAR gamma )-selective antagonists covalently bind to the PPAR gamma ligand binding domain with a unique binding mode.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
7RDX
 
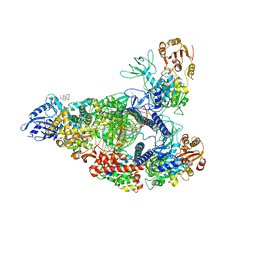 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - open class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1DMB
 
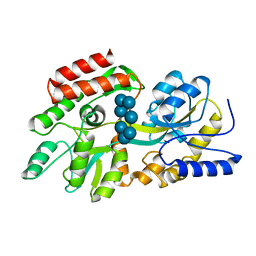 | |
1DOA
 
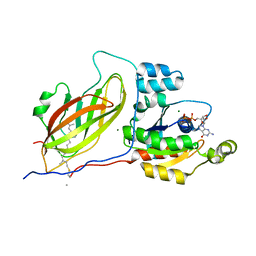 | | Structure of the rho family gtp-binding protein cdc42 in complex with the multifunctional regulator rhogdi | | Descriptor: | GERAN-8-YL GERAN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hoffman, G.R, Nassar, N, Cerione, R.C. | | Deposit date: | 1999-12-20 | | Release date: | 2000-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI.
Cell(Cambridge,Mass.), 100, 2000
|
|
6LOC
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 6-cyclobutyloxy-9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20014858 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
7RDZ
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - apo class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE3
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7D8X
 
 | | CryoEM structure of human gamma-secretase in complex with E2012 and L685458 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[(1S)-1-(4-fluorophenyl)ethyl]-3-[[3-methoxy-4-(4-methylimidazol-1-yl)phenyl]methylidene]piperidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
7RE1
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC (composite) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6LX8
 
 | | X-ray structure of human PPARalpha ligand binding domain-oleic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, OLEIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7RDY
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE2
 
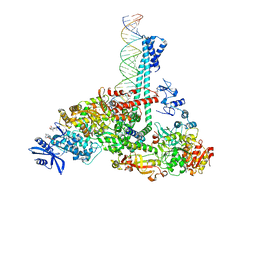 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(1)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1DL0
 
 | |
