4RTC
 
 | | Crystal structure of the green fluorescent variant, nowGFP, of the cyan Cerulean at pH 9.0 | | Descriptor: | GLYCEROL, nowGFP | | Authors: | Pletnev, V.Z, Pletneva, N.V, Pletnev, S.V. | | Deposit date: | 2014-11-14 | | Release date: | 2015-09-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the green fluorescent protein NowGFP with an anionic tryptophan-based chromophore.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RVW
 
 | |
4RW8
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in complex with (E)-3-(3-chloro-5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)phenyl)acrylonitrile (JLJ532), a non-nucleoside inhibitor' | | Descriptor: | (2E)-3-(3-chloro-5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, p51 subunit, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2014-12-01 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.878 Å) | | Cite: | Structure-Based Evaluation of Non-nucleoside Inhibitors with Improved Potency and Solubility That Target HIV Reverse Transcriptase Variants.
J.Med.Chem., 58, 2015
|
|
4RWU
 
 | | J-domain of Sis1 protein, Hsp40 co-chaperone from Saccharomyces cerevisiae | | Descriptor: | Protein SIS1 | | Authors: | Osipiuk, J, Zhou, M, Gu, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-05 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Roles of intramolecular and intermolecular interactions in functional regulation of the Hsp70 J-protein co-chaperone Sis1.
J.Mol.Biol., 427, 2015
|
|
4RX8
 
 | | SYK Catalytic Domain Complexed with a Potent Triazine Inhibitor2 | | Descriptor: | 3-{[(1R,2S)-2-aminocyclohexyl]amino}-5-(1H-indol-7-ylamino)-1,2,4-triazine-6-carboxamide, GLYCEROL, Tyrosine-protein kinase SYK | | Authors: | Lee, C.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery and profiling of a selective and efficacious syk inhibitor.
J.Med.Chem., 58, 2015
|
|
4RZF
 
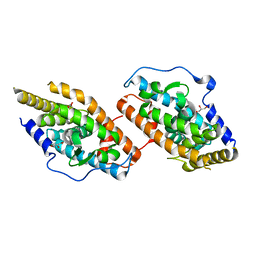 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, S441W mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
4RZC
 
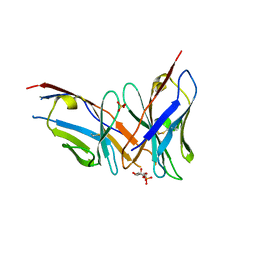 | | Fv M6P-1 in complex with mannose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, Fv M6P-1 heavy chain, Fv M6P-1 light chain, ... | | Authors: | Blackler, R.J, Evans, D.W, Evans, S.V, Muller-Loennies, S. | | Deposit date: | 2014-12-19 | | Release date: | 2015-11-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Single-chain antibody-fragment M6P-1 possesses a mannose 6-phosphate monosaccharide-specific binding pocket that distinguishes N-glycan phosphorylation in a branch-specific manner.
Glycobiology, 26, 2016
|
|
4S1P
 
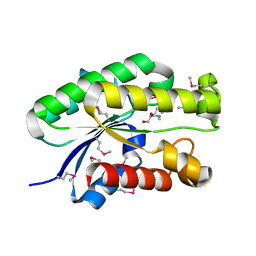 | | Shel_16390 protein, a putative SGNH hydrolase from Slackia heliotrinireducens | | Descriptor: | UNKNOWN LIGAND, Uncharacterized protein | | Authors: | Osipiuk, J, Cuff, M.E, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Shel_16390 protein, a putative SGNH hydrolase from Slackia heliotrinireducens
To be Published
|
|
4S29
 
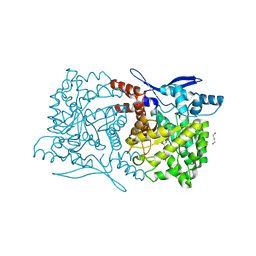 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide and Fe | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, FE (II) ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.382 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4TPP
 
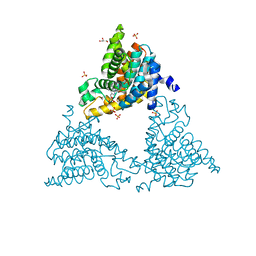 | | 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors | | Descriptor: | 1-[4-(3-{[1-(quinolin-2-yl)azetidin-3-yl]oxy}quinoxalin-2-yl)piperidin-1-yl]ethanone, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S. | | Deposit date: | 2014-06-09 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and preliminary biological evaluation of potent and selective 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors with improved solubility.
Bioorg.Med.Chem., 22, 2014
|
|
4RNF
 
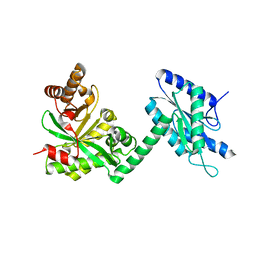 | |
4RP3
 
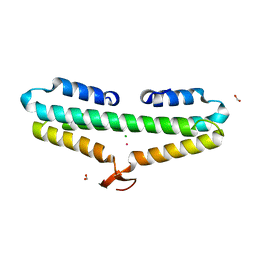 | | Crystal Structure of the L27 Domain of Discs Large 1 (target ID NYSGRC-010766) from Drosophila melanogaster bound to a potassium ion (space group P212121) | | Descriptor: | CHLORIDE ION, Disks large 1 tumor suppressor protein, FORMIC ACID, ... | | Authors: | Ghosh, A, Ramagopal, U, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-29 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of the L27 Domain of Disc Large Homologue 1 Protein Illustrate a Self-Assembly Module.
Biochemistry, 57, 2018
|
|
4RX7
 
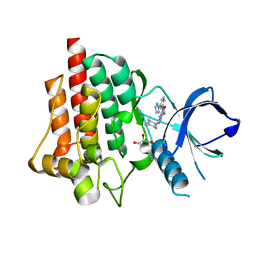 | | SYK Catalytic Domain Complexed with a Potent Triazine Inhibitor | | Descriptor: | 3-{[(1R,2S)-2-aminocyclohexyl]amino}-5-{[3-(2H-1,2,3-triazol-2-yl)phenyl]amino}-1,2,4-triazine-6-carboxamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Lee, C.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and profiling of a selective and efficacious syk inhibitor.
J.Med.Chem., 58, 2015
|
|
4RX9
 
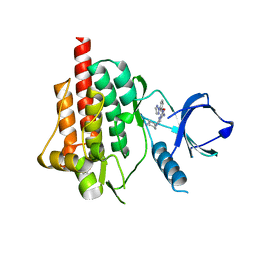 | | SYK Catalytic Domain Complexed with a Potent Pyrimidine Inhibitor | | Descriptor: | 2-{[(1R,2S)-2-aminocyclohexyl]amino}-4-{[3-(2H-1,2,3-triazol-2-yl)phenyl]amino}pyrimidine-5-carboxamide, GLYCEROL, Tyrosine-protein kinase SYK | | Authors: | Lee, C.C. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and profiling of a selective and efficacious syk inhibitor.
J.Med.Chem., 58, 2015
|
|
4S25
 
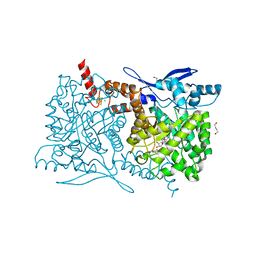 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide, S-adenosylhomocysteine, Fe4S4 cluster and Zn (trigonal crystal form) | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S2A
 
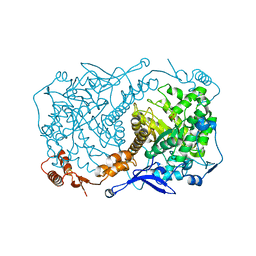 | | Crystal structure of Caulobacter crescentus ThiC with Fe4S4 cluster at remote site (holo form) | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, Phosphomethylpyrimidine synthase | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4RKS
 
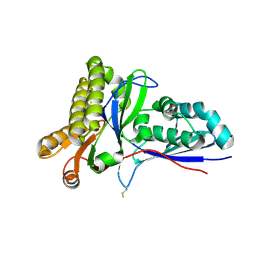 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (Mevalonate Bound) | | Descriptor: | (R)-MEVALONATE, ACETATE ION, GLYCEROL, ... | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RLU
 
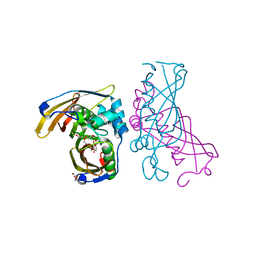 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with 2',4,4'-trihydroxychalcone | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 2',4,4'-TRIHYDROXYCHALCONE, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
4RNH
 
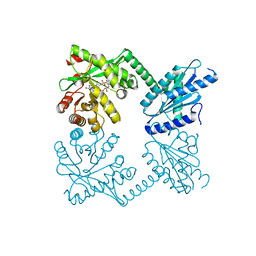 | | PaMorA tandem diguanylate cyclase - phosphodiesterase, c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Motility regulator | | Authors: | Phippen, C.W, Tews, I. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator.
Febs Lett., 588, 2014
|
|
4RO5
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM | | Descriptor: | GLYCEROL, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-27 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4RSX
 
 | |
4RUZ
 
 | |
4RW6
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) variant in complex with (E)-3-(3-chloro-5-(4-chloro-2-(2-(2,4-dioxo-3,4- dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)phenyl)acrylonitrile (JLJ494), a Non-nucleoside Inhibitor | | Descriptor: | (2E)-3-(3-chloro-5-{4-chloro-2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, p51 subunit, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2014-12-01 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Structure-Based Evaluation of Non-nucleoside Inhibitors with Improved Potency and Solubility That Target HIV Reverse Transcriptase Variants.
J.Med.Chem., 58, 2015
|
|
4RWK
 
 | | Crystal structure of V561M FGFR1 gatekeeper mutation (C488A, C584S, V561M) in complex with N-{3-[2-(3,5-DIMETHOXYPHENYL)ETHYL]-1H-PYRAZOL-5-YL}-4-[(3R,5S)-3,5-DIMETHYLPIPERAZIN-1-YL]BENZAMIDE (AZD4547) | | Descriptor: | Fibroblast growth factor receptor 1, MAGNESIUM ION, N-{3-[2-(3,5-dimethoxyphenyl)ethyl]-1H-pyrazol-5-yl}-4-[(3R,5S)-3,5-dimethylpiperazin-1-yl]benzamide | | Authors: | Sohl, C.D, Anderson, K.S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.982 Å) | | Cite: | Illuminating the Molecular Mechanisms of Tyrosine Kinase Inhibitor Resistance for the FGFR1 Gatekeeper Mutation: The Achilles' Heel of Targeted Therapy.
Acs Chem.Biol., 10, 2015
|
|
4RXX
 
 | | Crystal Structure of the N-terminal Domain of Human Ubiquitin Specific Protease 38 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Shen, L, Hu, J, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-12 | | Release date: | 2015-01-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the N-terminal Domain of Human Ubiquitin Specific Protease 38
to be published
|
|
