4P5Q
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-(2-chlorophenyl)-N-(3-ethoxypropyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-19 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
9FSV
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-22 | | Release date: | 2024-07-17 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
1V19
 
 | | 2-KETO-3-DEOXYGLUCONATE KINASE FROM THERMUS THERMOPHILUS | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-KETO-3-DEOXYGLUCONATE KINASE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2004-04-12 | | Release date: | 2004-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Thermus Thermophilus 2-Keto-3-Deoxygluconate Kinase: Evidence for Recognition of an Open Chain Substrate
J.Mol.Biol., 340, 2004
|
|
7KFR
 
 | |
9FST
 
 | | Yeast 20S proteasome with human beta1i (1-51) in complex with epoxyketone inhibitor LU-001i | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
To Be Published
|
|
2Q0N
 
 | | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Turnbull, A, Papagrigoriou, E, Pike, A.W, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide.
To be Published
|
|
5Y8Z
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-(3,6-dimethyl-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
6BO7
 
 | | Crystal structure of Plasmodium vivax hypoxanthine guanine phosphoribosyltransferase in complex with [3R,4R]-4-guanin-9-yl-3-((S)-2-hydroxy-2-phosphonoethyl)oxy-1-N-(phosphonopropionyl)pyrrolidine | | Descriptor: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | Authors: | Guddat, L.W, Keough, D.T, Rejman, D. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.856 Å) | | Cite: | Design of Plasmodium vivax Hypoxanthine-Guanine Phosphoribosyltransferase Inhibitors as Potential Antimalarial Therapeutics.
ACS Chem. Biol., 13, 2018
|
|
6C4J
 
 | | Ligand bound full length hUGDH with A104L substitution | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-1,2-PROPANEDIOL, ... | | Authors: | Beattie, N.R, Pioso, B.J, Wood, Z.A, Sidlo, A.M. | | Deposit date: | 2018-01-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Hysteresis and Allostery in Human UDP-Glucose Dehydrogenase Require a Flexible Protein Core.
Biochemistry, 57, 2018
|
|
4HBV
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-3-methyl-3,4-dihydroquinazolin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
6MAP
 
 | | F9 Pilus Adhesin FmlH Lectin Domain from E. coli UTI89 in Complex with Galactoside 5-nitro-2'-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-[1,1'-biphenyl]-3-carboxylic acid | | Descriptor: | 2'-(beta-D-galactopyranosyloxy)-5-nitro[1,1'-biphenyl]-3-carboxylic acid, Fimbrial adhesin FmlD, GLYCEROL, ... | | Authors: | Klein, R.D, Hultgren, S.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Biphenyl Gal and GalNAc FmlH Lectin Antagonists of Uropathogenic E. coli (UPEC): Optimization through Iterative Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
3R30
 
 | | MK2 kinase bound to Compound 2 | | Descriptor: | 1-(2-aminoethyl)-3-[2-(quinolin-3-yl)pyridin-4-yl]-1H-pyrazole-5-carboxylic acid, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, Fisher, M. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5ECO
 
 | | Crystal Structure of FIN219-FIP1 complex with JA, Leu and Mg | | Descriptor: | GLUTATHIONE, Glutathione S-transferase U20, Jasmonic acid-amido synthetase JAR1, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4JN9
 
 | | Crystal structure of the DepH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DepH, ... | | Authors: | Li, J, Wang, C, Zhang, Z.M, Zhou, J.H, Cheng, E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis.
Sci Rep, 4, 2014
|
|
7KJR
 
 | | Cryo-EM structure of SARS-CoV-2 ORF3a | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | Authors: | Kern, D.M, Hoel, C.M, Kotecha, A, Brohawn, S.G. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6MB1
 
 | |
2PW2
 
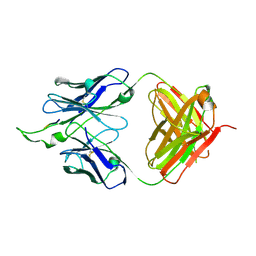 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKWKSL | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, peptide epitope | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2007-05-10 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications.
J.Virol., 83, 2009
|
|
5TNB
 
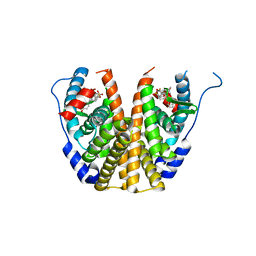 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S,L536S) in Complex with the OBHS-BSC, 4-bromophenyl (1R,2R,4S)-6-(4-(2-(dimethylamino)ethoxy)phenyl)-5-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Descriptor: | 4-bromophenyl (1S,2R,4S)-6-{4-[2-(dimethylamino)ethoxy]phenyl}-5-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor | | Authors: | Nwachukwu, J.C, Sharma, N, Carlson, K.E, Srinivasan, S, Sharma, A, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploring the Structural Compliancy versus Specificity of the Estrogen Receptor Using Isomeric Three-Dimensional Ligands.
ACS Chem. Biol., 12, 2017
|
|
4CS2
 
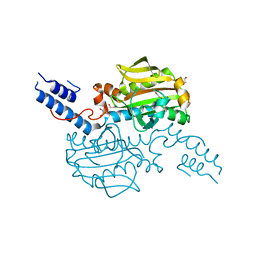 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in its apo form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PYRROLYSINE--TRNA LIGASE | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
8UTJ
 
 | | E. coli 70S ribosome with unmodified lys-tRNAPro(GGG) bound to slippery P-site CCC-C codon in the 0 frame | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-10-31 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
7X73
 
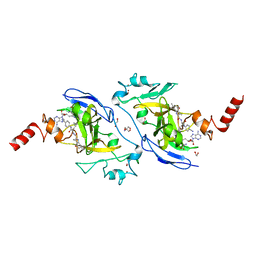 | | Structure of G9a in complex with RK-701 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A specific G9a inhibitor unveils BGLT3 lncRNA as a universal mediator of chemically induced fetal globin gene expression.
Nat Commun, 14, 2023
|
|
8URM
 
 | | E. coli 70S ribosome with unmodified tRNAPro(GGG) bound to slippery P-site CCC-C codon and tRNAVal(UAC) in the A site | | Descriptor: | 13S ribosomal RNA, 16S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-10-26 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
1VLO
 
 | |
8UXB
 
 | | E. coli 70S ribosome with unmodified P/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-11-09 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
7XMD
 
 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, the structure complexed with an allosteric inhibitor N4 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
