4MVL
 
 | |
4MNC
 
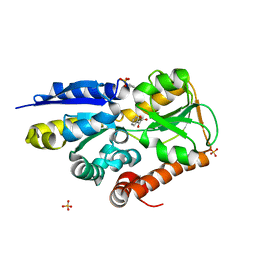 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P21 | | Descriptor: | BENZOYL-FORMIC ACID, SULFATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1HQ8
 
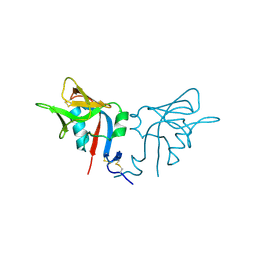 | | CRYSTAL STRUCTURE OF THE MURINE NK CELL-ACTIVATING RECEPTOR NKG2D AT 1.95 A | | Descriptor: | NKG2-D | | Authors: | Wolan, D.W, Teyton, L, Rudolph, M.G, Villmow, B, Bauer, S, Busch, D.H, Wilson, I.A. | | Deposit date: | 2000-12-14 | | Release date: | 2001-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A.
Nat.Immunol., 2, 2001
|
|
1HII
 
 | |
1HLU
 
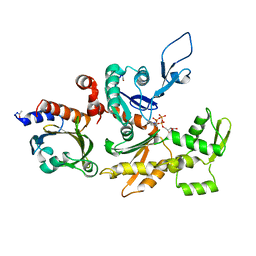 | | STRUCTURE OF BOVINE BETA-ACTIN-PROFILIN COMPLEX WITH ACTIN BOUND ATP PHOSPHATES SOLVENT ACCESSIBLE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BETA-ACTIN, CALCIUM ION, ... | | Authors: | Chik, J.K, Lindberg, U, Schutt, C.E. | | Deposit date: | 1997-05-30 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of an open state of beta-actin at 2.65 A resolution.
J.Mol.Biol., 263, 1996
|
|
5JPH
 
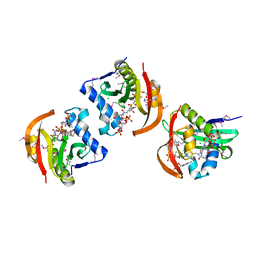 | | Structure of a GNAT acetyltransferase SACOL1063 from Staphylococcus aureus in complex with CoA | | Descriptor: | Acetyltransferase SACOL1063, CHLORIDE ION, COENZYME A | | Authors: | Majorek, K.A, Osinski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-06-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Insight into the 3D structure and substrate specificity of previously uncharacterized GNAT superfamily acetyltransferases from pathogenic bacteria.
Biochim.Biophys.Acta, 1865, 2016
|
|
4MLR
 
 | |
1HWH
 
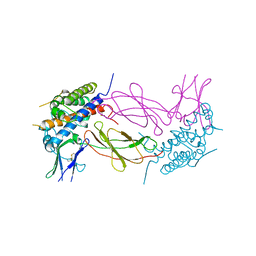 | |
1HNF
 
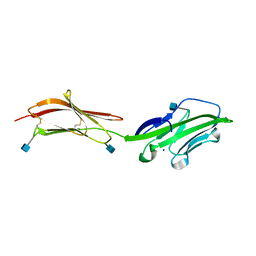 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR REGION OF THE HUMAN CELL ADHESION MOLECULE CD2 AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD2, SODIUM ION | | Authors: | Bodian, D.L, Jones, E.Y, Harlos, K, Stuart, D.I, Davis, S.J. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 A resolution.
Structure, 2, 1994
|
|
7U9J
 
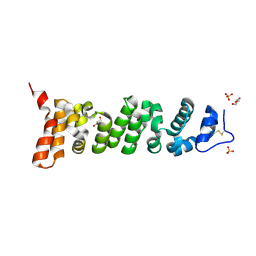 | | Crystal structure of Mesothelin-207 fragment | | Descriptor: | GLYCEROL, Isoform 3 of Mesothelin, SULFATE ION | | Authors: | Zhan, J, Esser, L, Lin, D, Tang, W.K, Xia, D. | | Deposit date: | 2022-03-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structures of Cancer Antigen Mesothelin and Its Complexes with Therapeutic Antibodies.
Cancer Res Commun, 3, 2023
|
|
1HSI
 
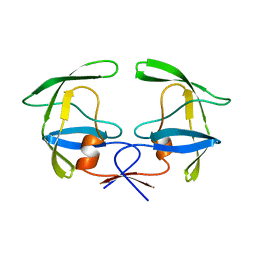 | |
7UED
 
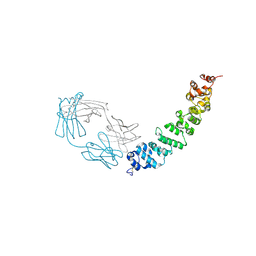 | | Crystal structure of full length mesothelin bound with MORAb-009 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Isoform 4 of Mesothelin, ... | | Authors: | Zhan, J, Esser, L, Lin, D, Tang, W.K, Xia, D. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of Cancer Antigen Mesothelin and Its Complexes with Therapeutic Antibodies.
Cancer Res Commun, 3, 2023
|
|
5JVG
 
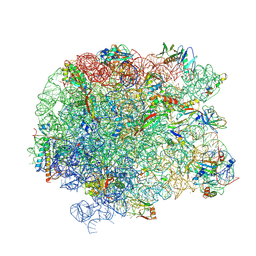 | | The large ribosomal subunit from Deinococcus radiodurans in complex with avilamycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Krupkin, M, Wekselman, I, Matzov, D, Eyal, Z, Diskin Posner, Y, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2016-05-11 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.428 Å) | | Cite: | Avilamycin and evernimicin induce structural changes in rProteins uL16 and CTC that enhance the inhibition of A-site tRNA binding.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1HXW
 
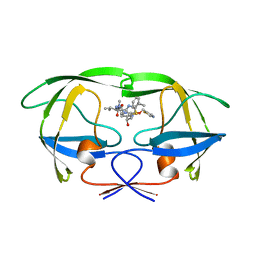 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-84538 | | Descriptor: | HIV-1 PROTEASE, RITONAVIR | | Authors: | Park, C.H, Nienaber, V, Kong, X.P. | | Deposit date: | 1997-01-24 | | Release date: | 1998-02-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1HUH
 
 | |
4N4K
 
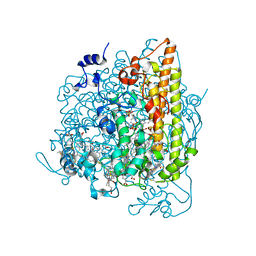 | | Kuenenia stuttgartiensis hydroxylamine oxidoreductase soaked in hydroxylamine | | Descriptor: | 1,2-ETHANEDIOL, HEME C, HYDROXYAMINE, ... | | Authors: | Maalcke, W.J, Dietl, A, Marritt, S.J, Butt, J.N, Jetten, M.S.M, Keltjens, J.T, Barends, T.R.M.B, Kartal, B. | | Deposit date: | 2013-10-08 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Biological NO Generation by Octaheme Oxidoreductases.
J.Biol.Chem., 289, 2014
|
|
1HQX
 
 | | R308K ARGINASE VARIANT | | Descriptor: | ARGINASE, MANGANESE (II) ION | | Authors: | Lavulo, L.T, Sossong Jr, T.M, Brigham-Burke, M.R, Doyle, M.L, Cox, J.D, Christianson, D.W, Ash, D.E. | | Deposit date: | 2000-12-20 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Subunit-subunit interactions in trimeric arginase. Generation of active monomers by mutation of a single amino acid.
J.Biol.Chem., 276, 2001
|
|
1HJN
 
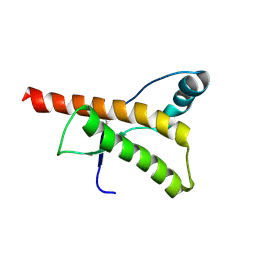 | | HUMAN PRION PROTEIN AT PH 7.0 | | Descriptor: | MAJOR PRION PROTEIN PRECURSOR | | Authors: | Calzolai, L, Zahn, R. | | Deposit date: | 2003-02-27 | | Release date: | 2003-07-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Influence of Ph on NMR Structure and Stability of the Human Prion Protein Globular Domain
J.Biol.Chem., 278, 2003
|
|
7UUJ
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin
To Be Published
|
|
4MMK
 
 | | Q8A Hfq from Pseudomonas aeruginosa | | Descriptor: | POTASSIUM ION, Protein hfq, SODIUM ION, ... | | Authors: | Murina, V.N, Filimonov, V.V, Melnik, B.S, Uhlein, M, Mueller, U, Weiss, M, Nikulin, A.D. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Effect of conserved intersubunit amino Acid substitutions on hfq protein structure and stability.
Biochemistry Mosc., 79, 2014
|
|
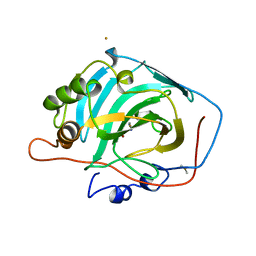
1HUG
 
 | |
1HIH
 
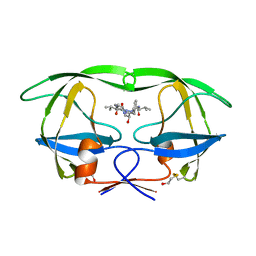 | |
4MVI
 
 | |
4MY5
 
 | | Crystal structure of the aromatic amino acid aminotransferase from Streptococcus mutants | | Descriptor: | Putative amino acid aminotransferase | | Authors: | Cong, X, Li, X, Ge, J, Feng, Y, Feng, X, Li, S. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of the aromatic-amino-acid aminotransferase from Streptococcus mutans.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5JS2
 
 | | Human Argonaute-2 Bound to a Modified siRNA | | Descriptor: | MAGNESIUM ION, PHENOL, PHOSPHATE ION, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2016-05-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural Analysis of Human Argonaute-2 Bound to a Modified siRNA Guide.
J.Am.Chem.Soc., 138, 2016
|
|
