1KE5
 
 | | CDK2 complexed with N-methyl-4-{[(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]amino}benzenesulfonamide | | Descriptor: | Cell division protein kinase 2, N-METHYL-4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
2K8P
 
 | | Characterisation of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation | | Descriptor: | Sclerostin | | Authors: | Veverka, V, Henry, A.J, Slocombe, P.M, Ventom, A, Mulloy, B, Muskett, F.W, Muzylak, M, Greenslade, K, Moore, A, Zhang, L, Gong, J, Qian, X, Paszty, C, Taylor, R.J, Robinson, M.K, Carr, M.D. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Structural Features and Interactions of Sclerostin: Molecular insight into a key regulator of Wnt-mediated bone formation
J.Biol.Chem., 284, 2009
|
|
2H24
 
 | | Crystal structure of human IL-10 | | Descriptor: | Interleukin-10 | | Authors: | Yoon, S.I, Walter, M.R. | | Deposit date: | 2006-05-18 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex.
J.Biol.Chem., 281, 2006
|
|
1URW
 
 | | CDK2 IN COMPLEX WITH AN IMIDAZO[1,2-b]PYRIDAZINE | | Descriptor: | 2-[4-(N-(3-DIMETHYLAMINOPROPYL)SULPHAMOYL)ANILINO]-, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Byth, K.F, Cooper, N, Culshaw, J.D, Heaton, D.W, Oakes, S.E, Minshull, C.A, Norman, R.A, Pauptit, R.A, Tucker, J.A, Breed, J, Pannifer, A, Rowsell, S, Stanway, J.J, Valentine, A.L, Thomas, A.P. | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Imidazo[1,2-B]Pyridazines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2KA4
 
 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | Descriptor: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
2HY6
 
 | | A seven-helix coiled coil | | Descriptor: | General control protein GCN4, HEXANE-1,6-DIOL | | Authors: | Liu, J, Zheng, Q, Deng, Y, Cheng, C.S, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A seven-helix coiled coil.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2K8X
 
 | |
2KJE
 
 | | NMR structure of CBP TAZ2 and adenoviral E1A complex | | Descriptor: | CREB-binding protein, Early E1A 32 kDa protein, ZINC ION | | Authors: | Ferreon, J.C, Martinez-Yamout, M, Dyson, H, Wright, P.E. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2K8F
 
 | | Structural Basis for the Regulation of p53 Function by p300 | | Descriptor: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | Authors: | Bai, Y, Feng, H, Jenkins, L.M, Durell, S.R, Wiodawer, A, Appella, E. | | Deposit date: | 2008-09-08 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for p300 Taz2-p53 TAD1 Binding and Modulation by Phosphorylation.
Structure, 17, 2009
|
|
7F80
 
 | |
7OGM
 
 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ)-Hfq | | Descriptor: | 3'ETS(LeuZ), Polyribonucleotide nucleotidyltransferase, RNA-binding protein Hfq | | Authors: | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
1OIR
 
 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | 1-(DIMETHYLAMINO)-3-(4-{{4-(2-METHYLIMIDAZO[1,2-A]PYRIDIN-3-YL)PYRIMIDIN-2-YL]AMINO}PHENOXY)PROPAN-2-OL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
6TSG
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG) in complex with TETRAC | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Gellrich, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | l-Thyroxin and the Nonclassical Thyroid Hormone TETRAC Are Potent Activators of PPAR gamma.
J.Med.Chem., 63, 2020
|
|
7ONB
 
 | | Structure of the U2 5' module of the A3'-SSA complex | | Descriptor: | MINX, PHD finger-like domain-containing protein 5A, RNU2, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
3EN2
 
 | | Three-dimensional structure of the protein priB from Ralstonia solanacearum at the resolution 2.3A. Northeast Structural Genomics Consortium target RsR213C. | | Descriptor: | POTASSIUM ION, Probable primosomal replication protein n | | Authors: | Kuzin, A.P, Neely, H, Wang, H, Sahdev, S, Foote, E.L, Xiao, R, Liu, J, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the protein priB from Ralstonia solanacearum at the resolution 2.3A. Northeast Structural Genomics Consortium target RsR213C.
To be Published
|
|
6WP7
 
 | |
4KO0
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH an anilinylpyrimidine derivative (JLJ-135) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-methoxypyrimidin-2-yl)amino]-2-[(3-methylbut-2-en-1-yl)oxy]benzonitrile, HIV-1 reverse transcriptase, ... | | Authors: | Das, K, Bauman, J.D, Arnold, E. | | Deposit date: | 2013-05-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Extension into the entrance channel of HIV-1 reverse transcriptase-Crystallography and enhanced solubility.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7SQB
 
 | | PPAR gamma LBD bound to Inverse Agonist SR10221 | | Descriptor: | (2S)-2-{5-[(5-{[(1S)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Frkic, R.L, Pederick, J.L, Bruning, J.B. | | Deposit date: | 2021-11-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PPAR gamma Corepression Involves Alternate Ligand Conformation and Inflation of H12 Ensembles.
Acs Chem.Biol., 18, 2023
|
|
7SQA
 
 | | PPAR gamma LBD bound to SR10221 and SMRT corepressor motif | | Descriptor: | (2S)-2-{5-[(5-{[(1S)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Frkic, R.L, Pederick, J.L, Bruning, J.B. | | Deposit date: | 2021-11-05 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | PPAR gamma Corepression Involves Alternate Ligand Conformation and Inflation of H12 Ensembles.
Acs Chem.Biol., 18, 2023
|
|
8GLU
 
 | |
7T3A
 
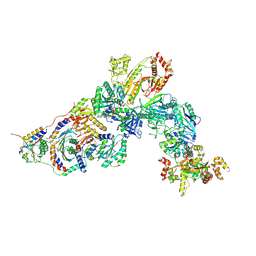 | | GATOR1-RAG-RAGULATOR - Inhibitory Complex | | Descriptor: | GATOR complex protein DEPDC5, GATOR complex protein NPRL2, GATOR complex protein NPRL3, ... | | Authors: | Egri, S.B, Shen, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
7T3C
 
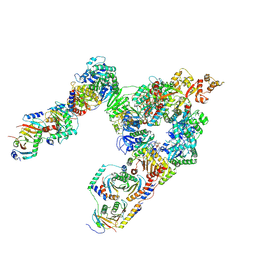 | | GATOR1-RAG-RAGULATOR - Dual Complex | | Descriptor: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | Authors: | Egri, S.B, Shen, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
7T3B
 
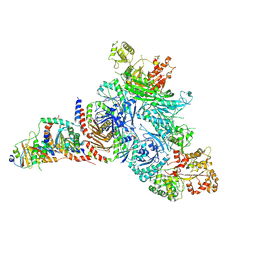 | | GATOR1-RAG-RAGULATOR - GAP Complex | | Descriptor: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | Authors: | Egri, S.B, Shen, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
1OIT
 
 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | 4-[(4-IMIDAZO[1,2-A]PYRIDIN-3-YLPYRIMIDIN-2-YL)AMINO]BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
7TRB
 
 | | CRYSTAL STRUCTURE OF FARNESOID X-ACTIVATED RECEPTOR COMPLEXED WITH COMPOUND-32 AKA (1S,3S)-N-({4-[5-(2-FLUOROPR OPAN-2-YL)-1,2,4-OXADIAZOL-3-YL]BICYCLO[2.2.2]OCTAN-1-YL}M ETHYL)-3-HYDROXY-N-[4'-(2-HYDROXYPROPAN-2-YL)-[1,1'-BIPHEN YL]-3-YL]-3-(TRIFLUOROMETHYL)CYCLOBUTANE-1-CARBOXAMIDE | | Descriptor: | (1s,3s)-N-({4-[5-(2-fluoropropan-2-yl)-1,2,4-oxadiazol-3-yl]bicyclo[2.2.2]octan-1-yl}methyl)-3-hydroxy-N-[4'-(2-hydroxypropan-2-yl)[1,1'-biphenyl]-3-yl]-3-(trifluoromethyl)cyclobutane-1-carboxamide, Bile acid receptor, co-activator | | Authors: | Khan, J.A, Ruzanov, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of BMS-986339, a Pharmacologically Differentiated Farnesoid X Receptor Agonist for the Treatment of Nonalcoholic Steatohepatitis.
J.Med.Chem., 65, 2022
|
|
