6RCY
 
 | |
6J1X
 
 | | WWP1 close conformation | | Descriptor: | GLYCEROL, NEDD4-like E3 ubiquitin-protein ligase WWP1 | | Authors: | Liu, Z.H. | | Deposit date: | 2018-12-30 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A multi-lock inhibitory mechanism for fine-tuning enzyme activities of the HECT family E3 ligases.
Nat Commun, 10, 2019
|
|
4QPL
 
 | | Crystal structure of RNF146(RING-WWE)/UbcH5a/iso-ADPr complex | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Wang, Z, DaRosa, P.A, Klevit, R.E, Xu, W. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal.
Nature, 517, 2015
|
|
6DHC
 
 | | X-ray structure of BACE1 in complex with a bicyclic isoxazoline carboxamide as the P3 ligand | | Descriptor: | (3R,3aR,6aS)-N-[(4R,7S,8S,10R,13S)-8-hydroxy-10,17-dimethyl-7-(2-methylpropyl)-5,11,14-trioxo-13-(propan-2-yl)-2-thia-6,12,15-triazaoctadecan-4-yl]hexahydrofuro[3,2-d][1,2]oxazole-3-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Mesecar, A.D, Lendy, E.K. | | Deposit date: | 2018-05-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design, synthesis, X-ray studies, and biological evaluation of novel BACE1 inhibitors with bicyclic isoxazoline carboxamides as the P3 ligand.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
8WDT
 
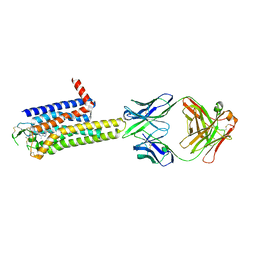 | | Crystal structure of the human adenosine A2A receptor in complex with photoresponsive ligand photoNECA(blue) | | Descriptor: | (2S,3S,4R,5R)-5-(6-amino-2-((E)-phenyldiazenyl)-9H-purin-9-yl)-N-ethyl-3,4-dihydroxytetrahydrofuran-2-carboxamide, Adenosine receptor A2a, Antibody Fab fragment heavy chain, ... | | Authors: | Araya, T, Asada, H, Iwata, S, Im, D.H. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Crystal structure reveals the binding mode and selectivity of a photoswitchable ligand for the adenosine A 2A receptor.
Biochem.Biophys.Res.Commun., 695, 2023
|
|
6XKC
 
 | | Crystal structure of E3 ligase | | Descriptor: | Protein fem-1 homolog C | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
2MGY
 
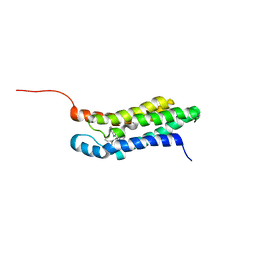 | | Solution structure of the mitochondrial translocator protein (TSPO) in complex with its high-affinity ligand PK11195 | | Descriptor: | N-[(2R)-butan-2-yl]-1-(2-chlorophenyl)-N-methylisoquinoline-3-carboxamide, Translocator protein | | Authors: | Jaremko, M, Jaremko, L, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the mitochondrial translocator protein in complex with a diagnostic ligand.
Science, 343, 2014
|
|
2ZMH
 
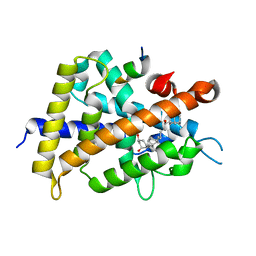 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1R,2E,4R)-4-hydroxy-1-methyl-4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]but-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-18 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
6IMJ
 
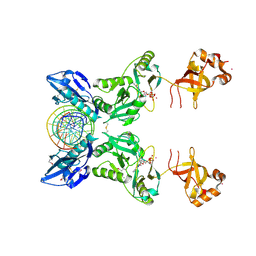 | | The crystal structure of Se-AsfvLIG:DNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
7NEC
 
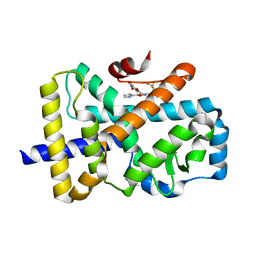 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM217 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-2-yl)-1,2-oxazol-4-yl]methoxy]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Somsen, B.A, de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
8R36
 
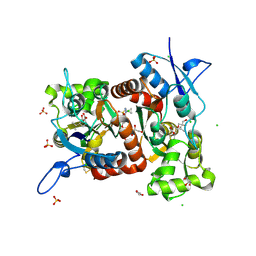 | | Crystal structure of the Gluk1 ligand-binding domain in complex with kainate and BPAM538 at 1.90 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-(3-methoxyphenoxy)-2,3-dihydro-1$l^{6},2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | Authors: | Bay, Y, Frantsen, S.M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2023-11-08 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GluK1 ligand-binding domain with kainate and the full-spanning positive allosteric modulator BPAM538.
J.Struct.Biol., 216, 2024
|
|
5OX2
 
 | |
6IMN
 
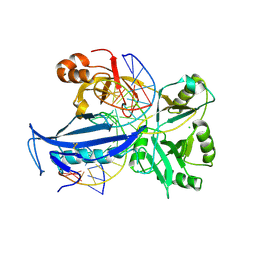 | | The crystal structure of AsfvLIG:CT2 complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*CP*CP*GP*CP*AP*TP*CP*CP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*GP*TP*CP*GP*GP*AP*CP*TP*GP*G)-3'), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
1I37
 
 | |
1I38
 
 | |
8R32
 
 | | Crystal structure of the GluK2 ligand-binding domain in complex with L-glutamate and BPAM344 at 1.60 A resolution | | Descriptor: | 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Bay, Y, Jeppesen, M.E, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2023-11-08 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The positive allosteric modulator BPAM344 and L-glutamate introduce an active-like structure of the ligand-binding domain of GluK2.
Febs Lett., 598, 2024
|
|
6AD9
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-9 | | Descriptor: | 12-mer peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, 3-[(1E)-1-{8-[(4-methyl-2-propyl-1H-benzimidazol-1-yl)methyl]dibenzo[b,e]oxepin-11(6H)-ylidene}ethyl]-1,2,4-oxadiazol-5(4H)-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Takahashi, Y, Suzuki, M, Yamamoto, K, Saito, J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Dihydrodibenzooxepine Peroxisome Proliferator-Activated Receptor (PPAR) Gamma Ligands of a Novel Binding Mode as Anticancer Agents: Effective Mimicry of Chiral Structures by Olefinic E/ Z-Isomers.
J. Med. Chem., 61, 2018
|
|
9EQK
 
 | |
5AIQ
 
 | | Crystal structure of ligand-free NadR | | Descriptor: | TRANSCRIPTIONAL REGULATOR, MARR FAMILY | | Authors: | Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2015-02-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Molecular Basis of Ligand-Dependent Regulation of Nadr, the Transcriptional Repressor of Meningococcal Virulence Factor Nada.
Plos Pathog., 12, 2016
|
|
8X6B
 
 | | Crystal structure of immune receptor PVRIG in complex with ligand Nectin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nectin-2, Transmembrane protein PVRIG | | Authors: | Hu, S.T, Han, P, Wang, H, Qi, J.X. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the immune recognition and selectivity of the immune receptor PVRIG for ligand Nectin-2.
Structure, 32, 2024
|
|
5C91
 
 | | NEDD4 HECT with covalently bound indole-based inhibitor | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, methyl (2E)-4-{[(5-methoxy-1,2-dimethyl-1H-indol-3-yl)carbonyl]amino}but-2-enoate | | Authors: | Span, I, Smith, A.T, Kathman, S, Statsyuk, A.V, Rosenzweig, A.C. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A Small Molecule That Switches a Ubiquitin Ligase From a Processive to a Distributive Enzymatic Mechanism.
J. Am. Chem. Soc., 137, 2015
|
|
4MBX
 
 | | Structure of unliganded B-Lymphotropic Polyomavirus VP1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Major Capsid Protein VP1 | | Authors: | Khan, Z.M, Neu, U, Schuch, B, Stehle, T. | | Deposit date: | 2013-08-21 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of B-Lymphotropic Polyomavirus VP1 in Complex with Oligosaccharide Ligands.
Plos Pathog., 9, 2013
|
|
3RPG
 
 | | Bmi1/Ring1b-UbcH5c complex structure | | Descriptor: | E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | Bentley, M.L, Dong, K.C, Cochran, A.G. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6485 Å) | | Cite: | Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex.
Embo J., 30, 2011
|
|
6IML
 
 | | The crystal structure of AsfvLIG:CT1 complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*CP*CP*GP*CP*AP*TP*CP*CP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*GP*GP*GP*AP*TP*GP*CP*GP*T)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*CP*TP*GP*G)-3'), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
3AUN
 
 | | Crystal structure of the rat vitamin D receptor ligand binding domain complexed with YR335 and a synthetic peptide containing the NR2 box of DRIP 205 | | Descriptor: | (2R)-2-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}butane-1,4-diol, DRIP 205 NR2 box peptide, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-02-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Design, synthesis and X-ray crystallographic study of new nonsecosteroidal vitamin D receptor ligands
Bioorg.Med.Chem.Lett., 21, 2011
|
|
